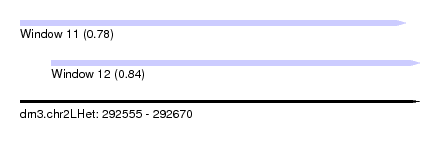
| Sequence ID | dm3.chr2LHet |
|---|---|
| Location | 292,555 – 292,670 |
| Length | 115 |
| Max. P | 0.837298 |

| Location | 292,555 – 292,666 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 60.70 |
| Shannon entropy | 0.62511 |
| G+C content | 0.43836 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -8.45 |
| Energy contribution | -9.13 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2LHet 292555 111 + 368872 -CGGAGGUGUAUA-AGCCCAACUGCGCAAAAUGUAAAUCGGCAUCGGAAAACUCCCAGCGGGGUAUUCGUUGUGCGAACAAAACGGAUGUCCGCCGUUAGCCU-CGAAAAUACA -..(((((.....-.(((....((((.....))))....)))...(((....))).(((((((((((((((.((....)).)))))))).)).))))).))))-)......... ( -31.30, z-score = -0.36, R) >droEre2.scaffold_4784 2363052 100 - 25762168 AAGAAAAA-UACA-AACCGACUUGUCCAAACUGUAA------UCCGGGAAACCGACAAAAGGAUGUU---UCUCUGAGCAACA---AGAUACAUCCGUAGAUUCAGAAGUUGCU ........-....-...((((((.......(((.((------(((((....)))......((((((.---(((.((.....))---))).))))))...))))))))))))).. ( -24.71, z-score = -2.65, R) >dp4.chr3 7785266 105 - 19779522 AAGAUACU-UACA-AACCGGUAUGUCCCAAAUGUAACCC---UCCGGGAAACCGGCAGAAGGAUGUG---UUAAACAACAACAAGCAGACACAUCCUUAGAAA-AAAGUUUGCA ........-..((-(((.(((((((.....)))).)))(---(((((....)))).))(((((((((---((...............))))))))))).....-...))))).. ( -28.96, z-score = -3.26, R) >droPer1.super_74 92554 107 - 351172 AAGAUACUGUAUGUAUCCGGUAUGUCCCAAAUGUAACCC---UCCGGGAAACCGGCAGAAUGAUGUG---UUAAACAGCAACAAUCAGACGCAUCCGUAGAAA-AAAGUUUGCA ..(((((.....))))).((..((((............(---(((((....)))).))..((((.((---((....))))...))))))))...))(((((..-....))))). ( -27.20, z-score = -2.01, R) >consensus AAGAAACU_UACA_AACCGAUAUGUCCAAAAUGUAACCC___UCCGGGAAACCGGCAGAAGGAUGUG___UUAAACAACAACAAGCAGACACAUCCGUAGAAA_AAAAUUUGCA ................................(((........((((....)))).....((((((........................))))))..............))). ( -8.45 = -9.13 + 0.69)

| Location | 292,564 – 292,670 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 60.06 |
| Shannon entropy | 0.64634 |
| G+C content | 0.44236 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -7.83 |
| Energy contribution | -8.77 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2LHet 292564 106 + 368872 AUA-AGCCCAACUGCGCAAAAUGUAAAUCGGCAUCGGAAAACUCCCAGCGGGGUAUUCGUUGUGCGAACAAAACGGAUGUCCGCCGUUAGCCUCGAAAAUACAAACA-- ...-.((.(....).))....((((..(((((...(((....))).(((((((((((((((.((....)).)))))))).)).))))).)))..))...))))....-- ( -25.90, z-score = -0.13, R) >droEre2.scaffold_4784 2363061 97 - 25762168 ACA-AACCGACUUGUCCAAACUGUAA------UCCGGGAAACCGACAAAAGGAUGUUUCUCUGAGCAACAA----GAUACAU-CCGUAGAUUCAGAAGUUGCUAAGUCA ...-....((((((..(((.(((.((------(((((....)))......((((((.(((.((.....)))----)).))))-))...)))))))...))).)))))). ( -29.80, z-score = -3.80, R) >dp4.chr3 7785275 100 - 19779522 ACA-AACCGGUAUGUCCCAAAUGUAACCC---UCCGGGAAACCGGCAGAAGGAUGUGUUAAACAACAACAAG-CAGACACAU-CCUUAGAA--AAAAGUUUGCAAGAA- .((-(((.(((((((.....)))).)))(---(((((....)))).))(((((((((((.............-..)))))))-))))....--....)))))......- ( -28.96, z-score = -3.48, R) >droPer1.super_74 92564 101 - 351172 AUGUAUCCGGUAUGUCCCAAAUGUAACCC---UCCGGGAAACCGGCAGAAUGAUGUGUUAAACAGCAACAAU-CAGACGCAU-CCGUAGAA--AAAAGUUUGCAAGAA- ........((..((((............(---(((((....)))).))..((((.((((....))))...))-))))))...-))(((((.--.....))))).....- ( -22.80, z-score = -1.17, R) >consensus ACA_AACCGAUAUGUCCAAAAUGUAACCC___UCCGGGAAACCGGCAGAAGGAUGUGUUAAACAGCAACAA__CAGACACAU_CCGUAGAA__AAAAGUUUCCAAGAA_ .................................((((....)))).....(((((((((................))))))).))........................ ( -7.83 = -8.77 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:04:49 2011