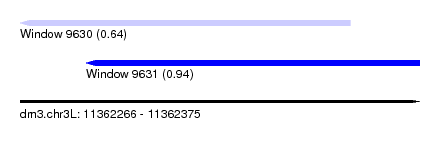
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,362,266 – 11,362,375 |
| Length | 109 |
| Max. P | 0.941298 |

| Location | 11,362,266 – 11,362,356 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 83.63 |
| Shannon entropy | 0.30800 |
| G+C content | 0.43567 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.99 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
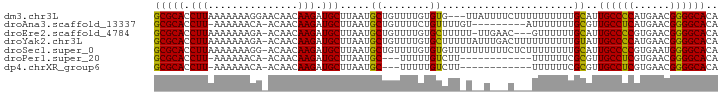
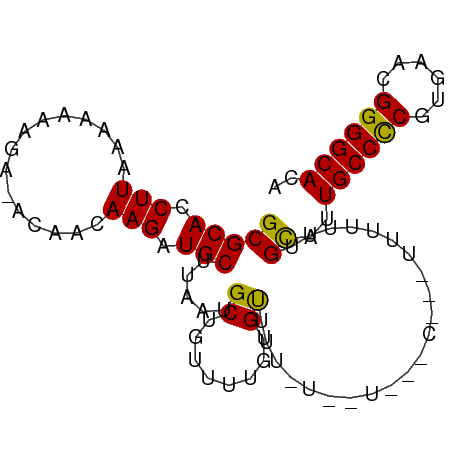
>dm3.chr3L 11362266 90 - 24543557 ACAACAAGAUGCUUAAUGCUGUUUUGUGUGUUAUUUU---CUUUUUUUUUUGCAUUGCCCCAUGAACGGGGCACACUUAUUGCCACGCAGUCG ((((((((((((.....)).))))))).)))......---...............((((((......)))))).....(((((...))))).. ( -20.20, z-score = -0.77, R) >droAna3.scaffold_13337 5644492 83 - 23293914 -CAACAAGAUGCUUAAUGCUGUUUUCUGUUUUGUAU---------UUUUUUGCGUUGCCUCAUGAACGGGGCACACUUAUUGCCACGCAGUCG -......(((((......((((((...((..((((.---------.....))))..)).....))))))((((.......))))..))).)). ( -17.20, z-score = 0.10, R) >droEre2.scaffold_4784 11366449 88 - 25762168 -CAACAAGAUGCUUAAUGCUGUUUUGUG----CUUUUUUUGAACGUUUUUUGCAUUGCCCCGUGAACGGGGCACACUUAUUGCCACGCAGUCG -..(((((((((.....)).)))))))(----(...........((.....))..(((((((....))))))).............))..... ( -23.90, z-score = -1.28, R) >droYak2.chr3L 11397547 92 - 24197627 -CAACAAGAUGCUUAAUGCUGUUUUGUGCUUUUUAUUUGACUUUUUUUUUUGUAUUGCCCCAUGAACGGGGCACACUUAUUGCCACGCAGUCG -..(((((((((.....)).)))))))...........(((..............((((((......)))))).......(((...)))))). ( -21.50, z-score = -1.66, R) >droSec1.super_0 3571584 92 - 21120651 -CAACAAGAUGCUUAAUGCUGUUUUGUGUGUUUUUUUUUUCUCUUUUUUUUGCAUUGCCCCGUGAAUGGGGCACACUUAUUGCCACGCAGUCG -..(((((((((.....)).)))))))............................(((((((....))))))).....(((((...))))).. ( -19.70, z-score = -0.54, R) >droPer1.super_20 1581831 77 + 1872136 -CAACAAGAUGCUUAAUGCU-UUUUGUC----UUU----------UUUUUCGCGUUGCCUCGUGAACGGGGCACACUUAUUGCCACGCAGUCG -..((((((.((.....)).-)))))).----...----------......(((((((((((....)))))))...........))))..... ( -22.30, z-score = -2.31, R) >dp4.chrXR_group6 2975718 77 + 13314419 -CAACAAGAUGCUUAAUGCU-UUUUGUC----UUU----------UUUUUCGCGUUGCCUCGUGAACGGGGCACACUUAUUGCCACGCAGUCG -..((((((.((.....)).-)))))).----...----------......(((((((((((....)))))))...........))))..... ( -22.30, z-score = -2.31, R) >consensus _CAACAAGAUGCUUAAUGCUGUUUUGUG__UUUUUUU________UUUUUUGCAUUGCCCCGUGAACGGGGCACACUUAUUGCCACGCAGUCG ...(((((((((.....)).)))))))............................((((((......)))))).....(((((...))))).. (-16.80 = -16.99 + 0.18)
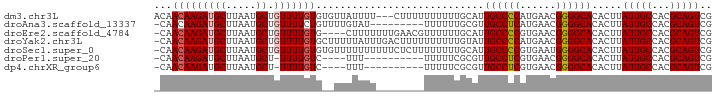
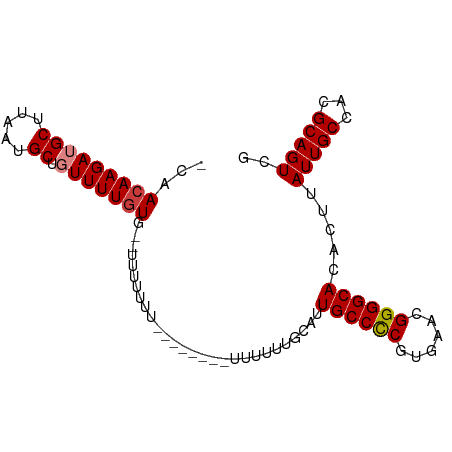
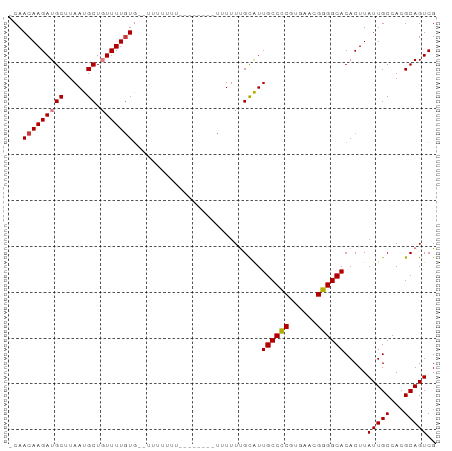
| Location | 11,362,284 – 11,362,375 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Shannon entropy | 0.34427 |
| G+C content | 0.40111 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -14.17 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11362284 91 - 24543557 GCGCACCUUAAAAAAAGGAACAACAAGAUGCUUAAUGCUGUUUUGUGUG---UUAUUUUCUUUUUUUUUUGCAUUGCCCCAUGAACGGGGCACA ..(((....((((((((((((((((((((((.....)).))))))).))---)....))))))))))..)))..((((((......)))))).. ( -28.50, z-score = -3.12, R) >droAna3.scaffold_13337 5644510 83 - 23293914 GCGCACCUU-AAAAAACA-ACAACAAGAUGCUUAAUGCUGUUUUCUGUUUUGU---------AUUUUUUUGCGUUGCCUCAUGAACGGGGCACA (((((....-((((.(((-(.((((..(.((........)).)..))))))))---------.))))..)))))((((((......)))))).. ( -20.90, z-score = -1.49, R) >droEre2.scaffold_4784 11366467 89 - 25762168 GCGCACCUUAAAAAAAGA-ACAACAAGAUGCUUAAUGCUGUUUUGUGCUUUUU-UUGAAC---GUUUUUUGCAUUGCCCCGUGAACGGGGCACA (((.((.((((((((((.-...(((((((((.....)).))))))).))))))-))))..---))....)))..(((((((....))))))).. ( -30.70, z-score = -3.63, R) >droYak2.chr3L 11397565 93 - 24197627 GCGCACCUUAAAAAAAGA-ACAACAAGAUGCUUAAUGCUGUUUUGUGCUUUUUAUUUGACUUUUUUUUUUGUAUUGCCCCAUGAACGGGGCACA ...........((((((.-...(((((((((.....)).))))))).)))))).....................((((((......)))))).. ( -22.00, z-score = -1.71, R) >droSec1.super_0 3571602 93 - 21120651 GCGCACCUUAAAAAAAGG-ACAACAAGAUGCUUAAUGCUGUUUUGUGUGUUUUUUUUUUCUCUUUUUUUUGCAUUGCCCCGUGAAUGGGGCACA ..(((....(((((((((-((((((((((((.....)).))))))).))))))))))))..........)))..(((((((....))))))).. ( -27.04, z-score = -2.38, R) >droPer1.super_20 1581849 77 + 1872136 GCGCACCUU-AAAAAACA-ACAACAAGAUGCUUAAUGC---UUUUUGUCUU------------UUUUUUCGCGUUGCCUCGUGAACGGGGCACA ((((.....-((((((..-...((((((.((.....))---.)))))).))------------))))...))))(((((((....))))))).. ( -22.30, z-score = -2.63, R) >dp4.chrXR_group6 2975736 77 + 13314419 GCGCACCUU-AAAAAACA-ACAACAAGAUGCUUAAUGC---UUUUUGUCUU------------UUUUUUCGCGUUGCCUCGUGAACGGGGCACA ((((.....-((((((..-...((((((.((.....))---.)))))).))------------))))...))))(((((((....))))))).. ( -22.30, z-score = -2.63, R) >consensus GCGCACCUUAAAAAAAGA_ACAACAAGAUGCUUAAUGCUGUUUUGUGUUUU_U__U___C___UUUUUUUGCAUUGCCCCGUGAACGGGGCACA (((((.(((...............))).))).....((........))......................))..((((((......)))))).. (-14.17 = -13.60 + -0.57)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:28 2011