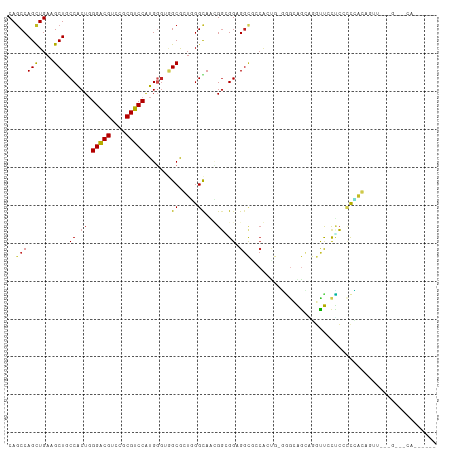
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,356,701 – 11,356,813 |
| Length | 112 |
| Max. P | 0.630937 |

| Location | 11,356,701 – 11,356,813 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 64.80 |
| Shannon entropy | 0.72440 |
| G+C content | 0.68519 |
| Mean single sequence MFE | -50.59 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.25 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11356701 112 + 24543557 CAGCCAGCUGAAGCUACCACUGGGUCGUCCGCGACCAUGGGUGGCGGUGGGUAAUGGCGGAGGCGCCACUG-GGGCAGCAGGUUCCUCCCCCACAGUUGCUGCUUCAGUGGCU ..(((((((.(.((((((....(((((....)))))...))))))..).)))..))))......(((((((-(((((((((.(...........).)))))))))))))))). ( -57.50, z-score = -1.95, R) >droSim1.chr3L 10746668 112 + 22553184 CAGCCAGCUGAAGCUACCACUGGGUCGUCCGCGACCAUGGGUGGCGGUGGGUAAUGGCGGAGGCGCCACUG-GGGCAGCAGGUUCCUCCCCCACACUUGCUGCUUCAGUGGCU ..(((((((.(.((((((....(((((....)))))...))))))..).)))..))))......(((((((-(((((((((((...........)))))))))))))))))). ( -61.40, z-score = -3.03, R) >droSec1.super_0 3566068 112 + 21120651 CAGCCAGCUGAAGCUACCACUGGGUCGUCCGCGACCAUGGGUGGCGGUGGGUAAUGGCGGAGGCGCCACUG-GGGCAGCAGGUUCCUCCCCCACACUUGCUGCUUCAGUGGCU ..(((((((.(.((((((....(((((....)))))...))))))..).)))..))))......(((((((-(((((((((((...........)))))))))))))))))). ( -61.40, z-score = -3.03, R) >droYak2.chr3L 11391468 106 + 24197627 CAGCCAGCUGAAGCUACCACUGGGUCGUCCGCGACCAUGGGAGGCGGUGGGCAACGGAGGAGGCGCCACUG-GAGCAGCUGGUUCCUCACCCACAGUUGAUGCUUCA------ .(((((((((..(((.((....(((((....)))))...)).)))....(....)((..((((.(((((((-...))).)))).))))..)).))))))..)))...------ ( -43.40, z-score = -0.24, R) >droEre2.scaffold_4784 11360748 112 + 25762168 CAGCCAGCUGAAGCUACCACUGGGUCGUCCGCGACCAUGGGAGGCGGUGGGCAAAGGGGGCGGCACCACUG-GGGCAGCUGGUUCCUCCCCCACAGUUUGUGCUUCAGAGGCA ..(((..(((((((....((((....((((((..((......))..))))))...(((((.((.(((((((-...))).)))).)).))))).))))....))))))).))). ( -54.30, z-score = -1.32, R) >droAna3.scaffold_13337 5639917 100 + 23293914 CAGCCAGCUGAAGCUGCCACUGGGACGUCCGCGUCCGUGGGAGGCGCUCGGCAGAGGAGGAG------CCG-AGCUGGUUUCCCCCUCCCCGGCGGAUUGCGAAGCA------ ..((..((.....(((((....(((((....)))))(.((((((.(((((((.........)------)))-))).((....))))))))))))))...))...)).------ ( -47.30, z-score = -0.67, R) >droGri2.scaffold_15110 14080975 98 + 24565398 CAGCCAGUUGAAGCUGCCACUGGGACGACCACGUCCAUGGCUGGCACUUGGUAGCGCUGGUGGCGCCGGUGGCGAGGAGGAUUCACUGGUUGCUGGAC--------------- ((((((((.((((((((((...(((((....))))).)))).))).(((.((..((((((.....)))))))).)))....)))))))))))......--------------- ( -42.00, z-score = -1.03, R) >droMoj3.scaffold_6680 6568804 95 - 24764193 CAGCCAGCUGAAGCUGCCGCUGGGACGCCCGCGUCCAUGCGUAGCACUCGGCAAGGGGGGCGGUGGCAGUACGGGCGGGGAGCCGCCCACCUGGU------------------ ..(((((.....(((((((((.(((((....))))).......((.(((.....)))..)))))))))))..((((((....))))))..)))))------------------ ( -50.60, z-score = -0.94, R) >droVir3.scaffold_13049 1657308 92 - 25233164 CAGCCAGCUGAAGCUGCCGCUGGGACGACCGCGUCCGUGUGUGGCGCUGGGCAGCGGGGGCGGCAACGG---AGGAGGUGAGCCGCCCACUUGGA------------------ ...(((((((.(((.(((((..(((((....)))))....))))))))...))))..(((((((.((..---.....))..)))))))...))).------------------ ( -46.20, z-score = -0.81, R) >droPer1.super_20 1577041 94 - 1872136 CAACCAGCUGAGGCUGCCGCUCGGACGUCCACGUCCAUGGGAUGCACUGGGCAGCGGCGGCGGCGGCG----CUGCAGUCGGCUGUGCUAUGGGGGCU--------------- (..(((((((..((((((((..(((((....)))))......(((.....)))))))))))..))))(----(.((((....))))))..)))..)..--------------- ( -44.90, z-score = -0.18, R) >dp4.chrXR_group6 2971098 97 - 13314419 CAGCCAGCUGAGGCUGCCGCUCGGACGUCCACGUCCAUGGGAUGCACUGGGCAGCGGCGGCGGCGGCGGCG-CUGGAGUCGGCUGUGCAAUGGGGGCU--------------- (((((.((((..((((((((..(((((....)))))......(((.....)))))))))))..))))(((.-.....)))))))).((.......)).--------------- ( -47.50, z-score = -0.06, R) >consensus CAGCCAGCUGAAGCUGCCACUGGGACGUCCGCGUCCAUGGGUGGCGCUGGGCAACGGCGGAGGCGCCACUG_GGGCAGCAGGUUCCUCCCCCACAGUU___G___CA______ ..((((((....))).((.((.(((((....))))).......((.....))...)).)).)))................................................. (-17.76 = -17.25 + -0.51)


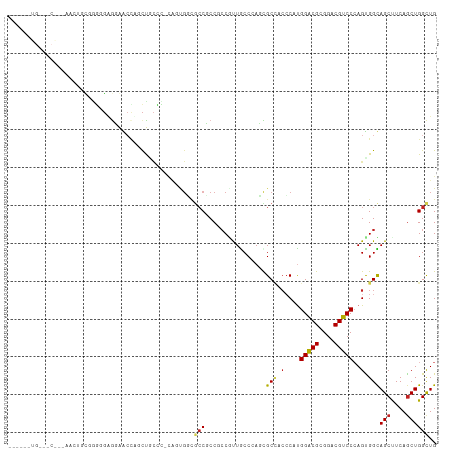
| Location | 11,356,701 – 11,356,813 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.80 |
| Shannon entropy | 0.72440 |
| G+C content | 0.68519 |
| Mean single sequence MFE | -44.67 |
| Consensus MFE | -14.67 |
| Energy contribution | -14.44 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11356701 112 - 24543557 AGCCACUGAAGCAGCAACUGUGGGGGAGGAACCUGCUGCCC-CAGUGGCGCCUCCGCCAUUACCCACCGCCACCCAUGGUCGCGGACGACCCAGUGGUAGCUUCAGCUGGCUG ((((((((((((.((.(((((((.(((((..((.((((...-))))))..))))).))).......((((.(((...))).))))......)))).)).))))))).))))). ( -55.40, z-score = -3.16, R) >droSim1.chr3L 10746668 112 - 22553184 AGCCACUGAAGCAGCAAGUGUGGGGGAGGAACCUGCUGCCC-CAGUGGCGCCUCCGCCAUUACCCACCGCCACCCAUGGUCGCGGACGACCCAGUGGUAGCUUCAGCUGGCUG ((((((((((((.....((((((.(((((..((.((((...-))))))..))))).))).))).....(((((....(((((....)))))..))))).))))))).))))). ( -55.20, z-score = -2.90, R) >droSec1.super_0 3566068 112 - 21120651 AGCCACUGAAGCAGCAAGUGUGGGGGAGGAACCUGCUGCCC-CAGUGGCGCCUCCGCCAUUACCCACCGCCACCCAUGGUCGCGGACGACCCAGUGGUAGCUUCAGCUGGCUG ((((((((((((.....((((((.(((((..((.((((...-))))))..))))).))).))).....(((((....(((((....)))))..))))).))))))).))))). ( -55.20, z-score = -2.90, R) >droYak2.chr3L 11391468 106 - 24197627 ------UGAAGCAUCAACUGUGGGUGAGGAACCAGCUGCUC-CAGUGGCGCCUCCUCCGUUGCCCACCGCCUCCCAUGGUCGCGGACGACCCAGUGGUAGCUUCAGCUGGCUG ------((((((((((.(((((((((((((....((..(..-..)..))...)))))....)))))((((..((...))..))))......))))))).))))))........ ( -40.90, z-score = -0.51, R) >droEre2.scaffold_4784 11360748 112 - 25762168 UGCCUCUGAAGCACAAACUGUGGGGGAGGAACCAGCUGCCC-CAGUGGUGCCGCCCCCUUUGCCCACCGCCUCCCAUGGUCGCGGACGACCCAGUGGUAGCUUCAGCUGGCUG .(((.(((((((....((((.(((((.((.((((.(((...-))))))).)).)))))........((((..((...))..))))......))))....)))))))..))).. ( -49.80, z-score = -1.75, R) >droAna3.scaffold_13337 5639917 100 - 23293914 ------UGCUUCGCAAUCCGCCGGGGAGGGGGAAACCAGCU-CGG------CUCCUCCUCUGCCGAGCGCCUCCCACGGACGCGGACGUCCCAGUGGCAGCUUCAGCUGGCUG ------......((...((((.(((((((((....)).(((-(((------(.........))))))).))))))..(((((....)))))).))))((((....)))))).. ( -50.70, z-score = -2.10, R) >droGri2.scaffold_15110 14080975 98 - 24565398 ---------------GUCCAGCAACCAGUGAAUCCUCCUCGCCACCGGCGCCACCAGCGCUACCAAGUGCCAGCCAUGGACGUGGUCGUCCCAGUGGCAGCUUCAACUGGCUG ---------------...((((...(((((((........(((...)))(((((..(((((....))))).......(((((....)))))..)))))...))).)))))))) ( -33.00, z-score = -0.72, R) >droMoj3.scaffold_6680 6568804 95 + 24764193 ------------------ACCAGGUGGGCGGCUCCCCGCCCGUACUGCCACCGCCCCCCUUGCCGAGUGCUACGCAUGGACGCGGGCGUCCCAGCGGCAGCUUCAGCUGGCUG ------------------.((((.(((((((....))))))...(((((.((((...((.(((..........))).))..))))((......)))))))....).))))... ( -39.60, z-score = 0.78, R) >droVir3.scaffold_13049 1657308 92 + 25233164 ------------------UCCAAGUGGGCGGCUCACCUCCU---CCGUUGCCGCCCCCGCUGCCCAGCGCCACACACGGACGCGGUCGUCCCAGCGGCAGCUUCAGCUGGCUG ------------------....((((((((((.........---.....)))))))((((((....(((((......)).)))((....))))))))((((....))))))). ( -38.04, z-score = -0.75, R) >droPer1.super_20 1577041 94 + 1872136 ---------------AGCCCCCAUAGCACAGCCGACUGCAG----CGCCGCCGCCGCCGCUGCCCAGUGCAUCCCAUGGACGUGGACGUCCGAGCGGCAGCCUCAGCUGGUUG ---------------.((.......)).((((((.(((.((----.((.((((((((.(((....))))).......(((((....)))))).))))).))))))).)))))) ( -38.00, z-score = -0.88, R) >dp4.chrXR_group6 2971098 97 + 13314419 ---------------AGCCCCCAUUGCACAGCCGACUCCAG-CGCCGCCGCCGCCGCCGCUGCCCAGUGCAUCCCAUGGACGUGGACGUCCGAGCGGCAGCCUCAGCUGGCUG ---------------.((.......)).((((((.((..((-.((.((((((...((.(((....))))).......(((((....)))))).))))).)))).)).)))))) ( -35.50, z-score = 0.28, R) >consensus ______UG___C___AACUGCGGGGGAGGAACCAGCUGCCC_CAGUGGCGCCGCCGCCGUUGCCCAGCGCCACCCAUGGACGCGGACGUCCCAGUGGCAGCUUCAGCUGGCUG .................................................(((................(((.(....(((((....)))))..).)))(((....)))))).. (-14.67 = -14.44 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:26 2011