| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,347,293 – 11,347,379 |
| Length | 86 |
| Max. P | 0.991476 |

| Location | 11,347,293 – 11,347,379 |
|---|---|
| Length | 86 |
| Sequences | 12 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.96 |
| Shannon entropy | 0.38459 |
| G+C content | 0.37655 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.39 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
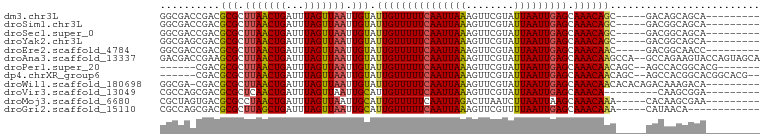
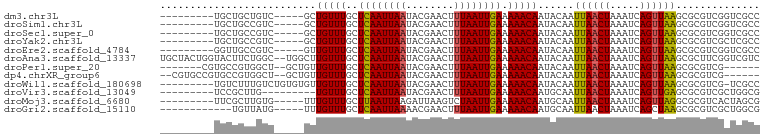
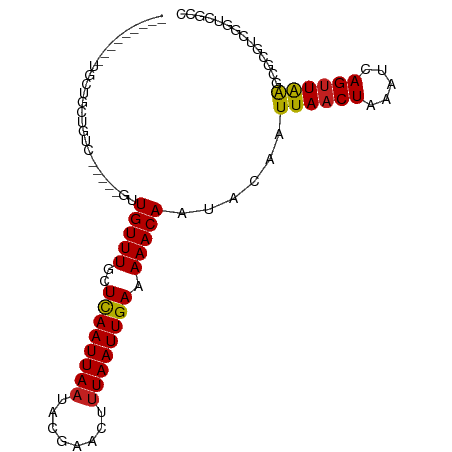
>dm3.chr3L 11347293 86 + 24543557 GGCGACCGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAGC-----GACAGCAGCA--------- .(((.....)))(((((((((...)))))).((((.(((((((((((((((........))))))))).)))))).-----.)))).))).--------- ( -18.80, z-score = -0.63, R) >droSim1.chr3L 10737034 86 + 22553184 GGCGACCGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAGC-----GACGGCAGCA--------- .((..(((.(((..(((((((...)))))))......((((((((((((((........))))))))).)))))))-----).)))..)).--------- ( -24.90, z-score = -2.50, R) >droSec1.super_0 3556709 86 + 21120651 GGCGACCGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAGC-----GACGGCAGCA--------- .((..(((.(((..(((((((...)))))))......((((((((((((((........))))))))).)))))))-----).)))..)).--------- ( -24.90, z-score = -2.50, R) >droYak2.chr3L 11381486 86 + 24197627 GGCGAGCGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAGC-----GACGGCAGCA--------- .....((..((((((((((((...)))))))......((((((((((((((........))))))))).)))))))-----).))...)).--------- ( -21.20, z-score = -0.77, R) >droEre2.scaffold_4784 11351291 86 + 25762168 GGCGACCGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAAC-----GACGGCAACC--------- ...(.(((..(((.(((((((...))))))).))).(((((((((((((((........))))))))).)))))).-----..))))....--------- ( -18.90, z-score = -1.22, R) >droAna3.scaffold_13337 5631050 98 + 23293914 GACGACCGAAGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAAGCCA--GCCAGAAGUACCAGUAGCA ............(((..((((..........(((..(((((((((((((((........))))))))).))))))..))--)..........))))))). ( -17.55, z-score = -0.80, R) >droPer1.super_20 1567224 85 - 1872136 ------CGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAACAGC--AGCCACGGCACG------- ------....((..(((((((...))))))).....(((((((((((((((........))))))))).))))))..))--.((....))...------- ( -17.40, z-score = -0.89, R) >dp4.chrXR_group6 2961190 90 - 13314419 ------CGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAACAGC--AGCCACGGCACGGCACG-- ------....(((((((((((...))))))......(((((((((((((((........))))))))).)))))).)))--.((....))...))...-- ( -19.40, z-score = -0.78, R) >droWil1.scaffold_180698 1760816 90 + 11422946 GGCGA-CGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAACACACAGACAAAGACA--------- .(((.-...)))..(((((((...))))))).(((.(((((((((((((((........))))))))).)))))).)))............--------- ( -17.60, z-score = -1.31, R) >droVir3.scaffold_13049 1638767 82 - 25233164 CGCCAGCGACGCGCUCAACUGAUUUAGUUAAUUGCAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACA---------CAAGCGGA--------- ((....)).(((((..(((((...)))))....))..((((((((((((((........))))))))).)))))---------...)))..--------- ( -16.30, z-score = -0.13, R) >droMoj3.scaffold_6680 6556081 86 - 24764193 CGCUAGUGACGCGCCUAACUGAUUUAGUUAAUUGCAUUGUUUUUCAAUUAGACUUAAUCUUAAUUAAGCAAACAAA-----CACAAGCGAA--------- ((((.(((..(((..((((((...))))))..))).((((((((.((((((........)))))).)).)))))).-----))).))))..--------- ( -18.70, z-score = -2.20, R) >droGri2.scaffold_15110 14069470 83 + 24565398 CGCCAGCGACGCGCUUAGCUGAUUUAGUUAAUUGCAUUGUUUUUCAAUUAAAGUUCGUUUUAAUUGAGCAAACAAA-----CAUAACA------------ ((....))..(((.(((((((...))))))).))).(((((((((((((((((....))))))))))).)))))).-----.......------------ ( -19.20, z-score = -1.74, R) >consensus GGCGACCGACGCGCUUAACUGAUUUAGUUAAUUGUAUUGUUUUUCAAUUAAAGUUCGUAUUAAUUGAGCAAACAAC_____GACAACAGCA_________ ..........(((.(((((((...))))))).))).(((((((((((((((........))))))))).))))))......................... (-14.59 = -14.39 + -0.20)
| Location | 11,347,293 – 11,347,379 |
|---|---|
| Length | 86 |
| Sequences | 12 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.96 |
| Shannon entropy | 0.38459 |
| G+C content | 0.37655 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -11.62 |
| Energy contribution | -11.56 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
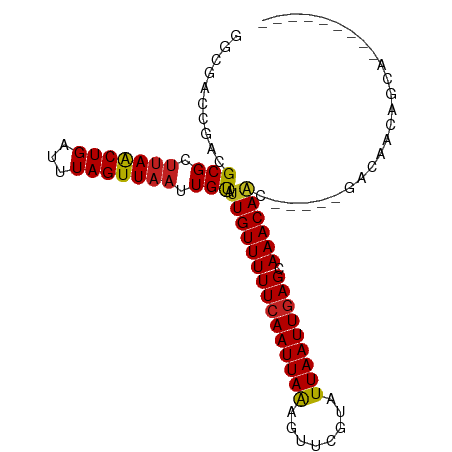
>dm3.chr3L 11347293 86 - 24543557 ---------UGCUGCUGUC-----GCUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCGGUCGCC ---------.((.((((.(-----(((((((..((((((((........)))))))).)))))......((((((.....))))))..))).)))).)). ( -24.70, z-score = -3.77, R) >droSim1.chr3L 10737034 86 - 22553184 ---------UGCUGCCGUC-----GCUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCGGUCGCC ---------.((.((((.(-----(((((((..((((((((........)))))))).)))))......((((((.....))))))..))).)))).)). ( -26.90, z-score = -4.55, R) >droSec1.super_0 3556709 86 - 21120651 ---------UGCUGCCGUC-----GCUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCGGUCGCC ---------.((.((((.(-----(((((((..((((((((........)))))))).)))))......((((((.....))))))..))).)))).)). ( -26.90, z-score = -4.55, R) >droYak2.chr3L 11381486 86 - 24197627 ---------UGCUGCCGUC-----GCUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCGCUCGCC ---------.((...((.(-----(((((((..((((((((........)))))))).)))))......((((((.....))))))..))).))...)). ( -20.20, z-score = -2.38, R) >droEre2.scaffold_4784 11351291 86 - 25762168 ---------GGUUGCCGUC-----GUUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCGGUCGCC ---------(((.((((.(-----(((((((..((((((((........)))))))).)))))......((((((.....))))))..))).)))).))) ( -25.50, z-score = -3.96, R) >droAna3.scaffold_13337 5631050 98 - 23293914 UGCUACUGGUACUUCUGGC--UGGCUUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCUUCGGUCGUC .(((((..(.....)..).--))))((((((..((((((((........)))))))).)))))).....((((((.....))))))((((....).))). ( -22.70, z-score = -2.72, R) >droPer1.super_20 1567224 85 + 1872136 -------CGUGCCGUGGCU--GCUGUUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCG------ -------(((((......(--(.((((((((..((((((((........)))))))).)))))))))).((((((.....)))))))))))...------ ( -22.70, z-score = -2.94, R) >dp4.chrXR_group6 2961190 90 + 13314419 --CGUGCCGUGCCGUGGCU--GCUGUUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCG------ --(((((...((....))(--(.((((((((..((((((((........)))))))).)))))))))).((((((.....)))))))))))...------ ( -24.20, z-score = -2.38, R) >droWil1.scaffold_180698 1760816 90 - 11422946 ---------UGUCUUUGUCUGUGUGUUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCG-UCGCC ---------............((((((((((..((((((((........)))))))).)))))))))).((((((.....))))))..(((...-.))). ( -22.70, z-score = -3.48, R) >droVir3.scaffold_13049 1638767 82 + 25233164 ---------UCCGCUUG---------UGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUGCAAUUAACUAAAUCAGUUGAGCGCGUCGCUGGCG ---------..((((.(---------(((((..((((((((........)))))))).)))).((((..((((((.....))))))..)))).)).)))) ( -21.40, z-score = -2.41, R) >droMoj3.scaffold_6680 6556081 86 + 24764193 ---------UUCGCUUGUG-----UUUGUUUGCUUAAUUAAGAUUAAGUCUAAUUGAAAAACAAUGCAAUUAACUAAAUCAGUUAGGCGCGUCACUAGCG ---------..((((.(((-----..(((..((((((((..((((.(((.((((((..........))))))))).))))))))))))))).))).)))) ( -22.00, z-score = -2.60, R) >droGri2.scaffold_15110 14069470 83 - 24565398 ------------UGUUAUG-----UUUGUUUGCUCAAUUAAAACGAACUUUAAUUGAAAAACAAUGCAAUUAACUAAAUCAGCUAAGCGCGUCGCUGGCG ------------.((((((-----(((((((..(((((((((......))))))))).)))))).)))..)))).......((((.(((...))))))). ( -20.50, z-score = -2.38, R) >consensus _________UGCUGCUGUC_____GUUGUUUGCUCAAUUAAUACGAACUUUAAUUGAAAAACAAUACAAUUAACUAAAUCAGUUAAGCGCGUCGGUCGCC ..........................(((((..((((((((........)))))))).)))))......((((((.....)))))).............. (-11.62 = -11.56 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:24 2011