| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,303,674 – 11,303,770 |
| Length | 96 |
| Max. P | 0.594032 |

| Location | 11,303,674 – 11,303,770 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.98 |
| Shannon entropy | 0.57217 |
| G+C content | 0.46513 |
| Mean single sequence MFE | -18.26 |
| Consensus MFE | -9.38 |
| Energy contribution | -9.91 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
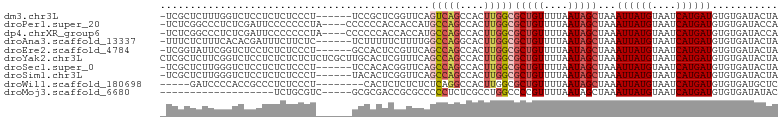
>dm3.chr3L 11303674 96 + 24543557 -UCGCUCUUUGGUCUCCUCUCUCCCU------UCCGCUCGGUUCAGUCAGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACUA -((((.(...................------...(((.((..((((..(((....)))))))..))...)))...((((((....))))))).))))..... ( -16.00, z-score = 0.18, R) >droPer1.super_20 1517137 98 - 1872136 -UCUCGGCCCUCUCGAUUCCCCCCCUA----CCCCCCACCACCAUGCCAGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACCA -..((((.....))))...........----.....(((((.((((...(((....)))(((((....))))).............)))).)).)))...... ( -17.40, z-score = -1.37, R) >dp4.chrXR_group6 2911303 98 - 13314419 -UCUCGGCCCUCUCGAUUCCCCCCCUA----CCCCCCACCACCAUGCCAGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACCA -..((((.....))))...........----.....(((((.((((...(((....)))(((((....))))).............)))).)).)))...... ( -17.40, z-score = -1.37, R) >droAna3.scaffold_13337 5587561 96 + 23293914 -UUUCUCUUUCACACGAUUUCUUCUC------UCUUUUUCUUUUGGCCAGGCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACUA -........((((((...........------.............(((((....)))))(((((....)))))...((((((....))))))))))))..... ( -21.70, z-score = -3.24, R) >droEre2.scaffold_4784 11307472 96 + 25762168 -UCGGUAUUCGGUCUCCUCUCUCCCU------GCCACUCCGUUCAGCCAGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACUA -..((((...((..........)).)------))).....((((((((((....)))))(((((....)))))...((((((....))))))...))).)).. ( -17.50, z-score = 0.01, R) >droYak2.chr3L 11339005 103 + 24197627 CUCGCUCUUCGGUCUCCUCUCUCUCUCUCGCUUGCACUCGUUUCAGCCAGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACUA ...............................(..(((........(((((....)))))(((((....)))))...((((((....)))))))))..)..... ( -17.60, z-score = -0.14, R) >droSec1.super_0 3516615 96 + 21120651 -UCGCUCUUGGGUCUCCUCUCUCCCU------UCCACACGGUUCAGCCAGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACUA -........(((..........))).------..(((((((.....)).(((....)))(((((....)))))...((((((....)))))))))))...... ( -22.90, z-score = -1.60, R) >droSim1.chr3L 10692575 96 + 22553184 -UCGCUCUUGGGUCUCCUCUCUCCCU------UACACUCGGUUCAGCCAGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACUA -........(((..........)))(------(((((..((.....)).(((....)))(((((....)))))...((((((....))))))))))))..... ( -21.40, z-score = -1.16, R) >droWil1.scaffold_180698 9719192 90 - 11422946 -----GAUCCCCACCGCCCUCUCCCU--------CACUCUCUCUCUCAGGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUGCUC -----....................(--------(((.(..........(((....)))(((((....)))))...((((((....))))))).))))..... ( -15.10, z-score = -0.73, R) >droMoj3.scaffold_6680 6515533 79 - 24764193 -------------------UCUGCGUC-----GCGCGACCGCGCCCCCUCUCGCCUGGCCCCGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUAUAC -------------------.....(((-----(((((((.(.(((...........))).).)))...........((((((....))))))))))))).... ( -15.60, z-score = -0.17, R) >consensus _UCGCUCUUCGGUCUCCUCUCUCCCU______UCCACUCCGUUCAGCCAGCCACUUGGCGCUGUUUUAAUAGCUAAAUUAUGUAAUCAUGAUGUGUGAUACUA .............................................(((((....)))))(((((....)))))...((((((....))))))........... ( -9.38 = -9.91 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:19 2011