| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,295,102 – 11,295,203 |
| Length | 101 |
| Max. P | 0.599615 |

| Location | 11,295,102 – 11,295,203 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Shannon entropy | 0.46831 |
| G+C content | 0.50947 |
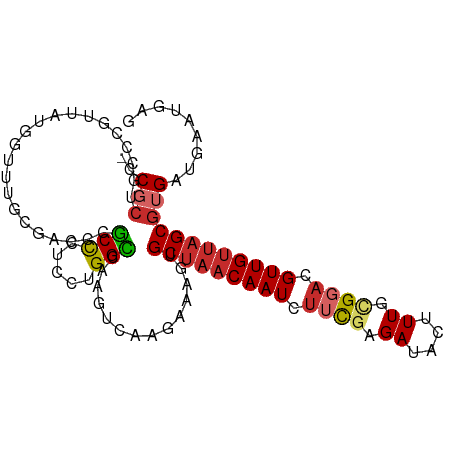
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
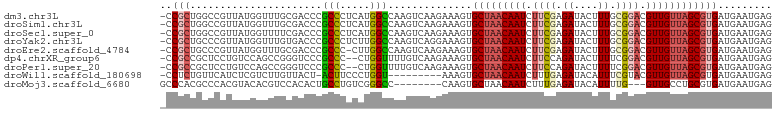
>dm3.chr3L 11295102 101 + 24543557 -CCGCUGGCCGUUAUGGUUUGCGACCCGCCCUCAUGGCCAAGUCAAGAAAGUGCUAACAAUCUUCGAGAUACUUUGCGGACGUUGUUAGCGUGAUGAAUGAG -.((((((((((...((...(((...)))))..)))))))............(((((((((.((((..(....)..)))).))))))))))))......... ( -30.70, z-score = -1.42, R) >droSim1.chr3L 10683667 101 + 22553184 -CCGCUGGCCGUUAUGGUUUGCGACCCGCCCUCAUGGCCAAGUCAAGAAAGUGCUAACAAUCUUCGAGAUACUUUGCGGACGUUGUUAGCGUGAUGAAUGAG -.((((((((((...((...(((...)))))..)))))))............(((((((((.((((..(....)..)))).))))))))))))......... ( -30.70, z-score = -1.42, R) >droSec1.super_0 3508216 101 + 21120651 -CCGCUGGCCGUUAUGGUUUUCGACCCGCCCUCAUGGCCAAGUCAAGAAAGUGCUAACAAUCUUCGAGAUACUUUGCGGACGUUGUUAGCGUGAUGAAUGAG -.((((((((((...(((.........)))...)))))))............(((((((((.((((..(....)..)))).))))))))))))......... ( -29.60, z-score = -1.32, R) >droYak2.chr3L 11330022 101 + 24197627 -CCGCUGCCCGUUAUGGUUUGUGACCCGCCCUCUUGGCCAAGUCAGGAAAGUGCUAACAAUCUUCGAGAUACUUUGCGGACGUUGUUAGCGUGAUGAAUGAG -........((((((..(((.((((..(((.....)))...)))).)))...(((((((((.((((..(....)..)))).)))))))))))))))...... ( -28.50, z-score = -1.04, R) >droEre2.scaffold_4784 11298814 100 + 25762168 -CCGCUGCCCGUUAUGGUUUGCGACCCGCCC-CUUGGCCAAGUCAAGAAAGUGCUAACAAUCUUCGAGAUACUUUGCGGACGUUGUUAGCGUGAUGAAUGAG -.(((.(((......)))..)))...(((..-((((((...)))))).....(((((((((.((((..(....)..)))).))))))))))))......... ( -28.30, z-score = -1.46, R) >dp4.chrXR_group6 2902291 99 - 13314419 -CCGCCGCUCCUGUCCAGCCGGGUCCCGCCC--CUGGUUUUGUCAAGAAAGUGCUAACAAUCUUCCAGAUACUUUUCGGACGUUGUUAGCGUGAUGAAUGAG -.((.(((((.((.(.(((((((.......)--))))))..).)).))....(((((((((..(((.((......))))).)))))))))))).))...... ( -27.00, z-score = -1.09, R) >droPer1.super_20 1508024 99 - 1872136 -CCGCCGCUCCUGUCCAGCCGGGUCCCGCCC--CUGGUUUUGUCAAGAAAGUGCUAACAAUCUUCCAGAUACUUUUCGGACGUUGUUAGCGUGAUGAAUGAG -.((.(((((.((.(.(((((((.......)--))))))..).)).))....(((((((((..(((.((......))))).)))))))))))).))...... ( -27.00, z-score = -1.09, R) >droWil1.scaffold_180698 1693481 91 + 11422946 -CCUCUGUUCAUCUCGUCUUGUUACU-ACUUCCCUGGU---------AAAGUGCUAACAAUCUUUGAGAUACAUUUCGUACGUUGUUAGCGUGAUGAAUGAG -.(((.(((((((.(......(((((-(......))))---------))...(((((((((...(((((....)))))...)))))))))).)))))))))) ( -26.90, z-score = -4.16, R) >droMoj3.scaffold_6680 18259253 91 + 24764193 GCCCACGCCCACGUACACGUCCACACUGCCUGUCGGGCC--------CAAGUGCUAACAAUCUUUGAGAUACAUUUUG---GUUGCCUGCGUGAUGAAUGAG ...(((((....((((..(((((((.....))).)))).--------...))))...(((((...((......))..)---))))...)))))......... ( -18.60, z-score = 1.12, R) >consensus _CCGCUGCCCGUUAUGGUUUGCGACCCGCCCUCCUGGCCAAGUCAAGAAAGUGCUAACAAUCUUCGAGAUACUUUGCGGACGUUGUUAGCGUGAUGAAUGAG ..(((......................(((.....)))..............(((((((((.((((.((....)).)))).))))))))))))......... (-15.16 = -15.72 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:14 2011