| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,293,132 – 11,293,235 |
| Length | 103 |
| Max. P | 0.832892 |

| Location | 11,293,132 – 11,293,235 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Shannon entropy | 0.40582 |
| G+C content | 0.48606 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -15.69 |
| Energy contribution | -15.44 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
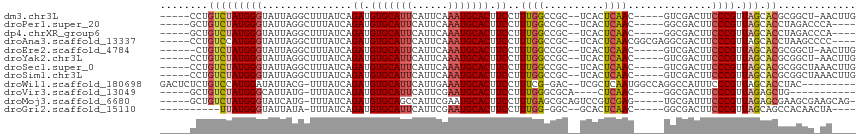
>dm3.chr3L 11293132 103 + 24543557 CAAGUU-AGCCGCGUGCUAACGGGAAGUCGAC-----GUUGAGUGA--GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUAGACAGG----- ...(((-(((.....))))))(((..((..((-----.....))..--))(((....(((.((((((((....)))))))).)))......))).....))).........----- ( -30.80, z-score = -2.33, R) >droPer1.super_20 1505985 100 - 1872136 ----UGGGUCUAGGUGCUAACGGGAAGUCGCC-----GUUGAGUGA--GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUAGACAGC----- ----(((((.(((((..((.((((.....(((-----(((.....)--)))))........((((((((....)))))))).)))).))..)))))..)))))........----- ( -34.60, z-score = -3.00, R) >dp4.chrXR_group6 2900253 100 - 13314419 ----UGGGUCUAGGUGCUAACGGGAAGUCGCC-----GUUGAGUGA--GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUAGACAGC----- ----(((((.(((((..((.((((.....(((-----(((.....)--)))))........((((((((....)))))))).)))).))..)))))..)))))........----- ( -34.60, z-score = -3.00, R) >droAna3.scaffold_13337 5577601 105 + 23293914 ----GGGGCUUAGGUGCUAACGGGAAGUCGCCUCGCCGUUGAGUGA--GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUGGACAGG----- ----(((..((((((..((.((((..(((((.((((......))))--)))))........((((((((....)))))))).)))).))..))))))..))).........----- ( -38.30, z-score = -2.43, R) >droEre2.scaffold_4784 11296912 102 + 25762168 CAAGUU-AGCCGCGUGCUAACGGGAAGUCGAC-----GUUGAGUGA--GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUAGACAG------ ...(((-(((.....))))))(((..((..((-----.....))..--))(((....(((.((((((((....)))))))).)))......))).....)))........------ ( -30.80, z-score = -2.64, R) >droYak2.chr3L 11328009 103 + 24197627 CAAGUU-AGCCGCGUGCUAACGGGAAGUCGAC-----GUUGAGUGA--GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUAGACAGG----- ...(((-(((.....))))))(((..((..((-----.....))..--))(((....(((.((((((((....)))))))).)))......))).....))).........----- ( -30.80, z-score = -2.33, R) >droSec1.super_0 3506202 104 + 21120651 CAAGUUUAGCCGCGUGCUAACGGGAAGUCGAC-----GUUGAGUGA--GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUAGACAGG----- ...((((((((((..(((((((.........)-----))).)))..--)))))....(((.((((((((....)))))))).)))..................)))))...----- ( -31.20, z-score = -2.35, R) >droSim1.chr3L 10681664 104 + 22553184 CAAGUUUAGCCGCGUGCUAACGGGAAGUCGAC-----GUUGAGUGA--GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUAGACAGG----- ...((((((((((..(((((((.........)-----))).)))..--)))))....(((.((((((((....)))))))).)))..................)))))...----- ( -31.20, z-score = -2.35, R) >droWil1.scaffold_180698 9744984 103 - 11422946 ---------GUAGGUGCUAACGGGAAAUGGCCUGGCCAUUGAGCGA--GUC-CGAAAGGAAGUGCAUUUCAAUGAAUGCACAUCUGAUAAA-CGUAAUAUCCAUGGACAGAGAGUC ---------....((((...((((......))))..(((((((((.--.((-(....)))..)))...))))))...)))).((((.....-(((.......)))..))))..... ( -28.40, z-score = -1.44, R) >droVir3.scaffold_13049 1582870 90 - 25233164 -----------CAGCUCUAACGGGAAGUCGCC-----GUUGAG----UGCGCCCAAAGGAAGUGCAUUCGAAUGAAUGCACAUCUGAUAAA-CAUAAUGCCCAUAGACAGC----- -----------..((((.((((((....).))-----))))))----).........(((.((((((((....)))))))).)))......-...................----- ( -24.20, z-score = -1.36, R) >droMoj3.scaffold_6680 6503479 104 - 24764193 -CUGCUUCGCUUCGCUCUAACGGGAAAUCGCA-----CUCGACGGACUGCGCUCAAAGGAAGUGCAUUCGAAUGGCUGCACAUCUGAUAAA-CAUGAUACCCAUAGACAGC----- -.......(((....((((..(((....((((-----.((....)).))))......(((.(((((..(....)..))))).)))......-.......))).)))).)))----- ( -21.70, z-score = 0.84, R) >droGri2.scaffold_15110 14020347 93 + 24565398 ----UAGUUGUGGCUGCUAACGGGAAGUCGCC-----GUUGAGUGC--GCC-CCAAAGGAAGUGCAUUCGAAUGAAUGCACAUCUGAUAAA-UAUAAUACCCAUAA---------- ----...(((.(((((((((((((....).))-----))).)))).--)))-.))).(((.((((((((....)))))))).)))......-..............---------- ( -28.20, z-score = -2.48, R) >consensus ____UU_AGCUGCGUGCUAACGGGAAGUCGCC_____GUUGAGUGA__GCGGCCAAAGGAAGUGCAUUUGAAUGAAUGCACAUCUGAUAAAGCCUAAUACCCAUAGACAGG_____ .....................(((...(((.........)))...............(((.((((((((....)))))))).)))..............))).............. (-15.69 = -15.44 + -0.25)

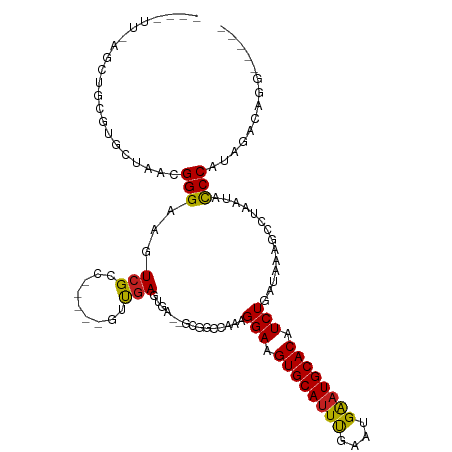
| Location | 11,293,132 – 11,293,235 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.61 |
| Shannon entropy | 0.40582 |
| G+C content | 0.48606 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -12.30 |
| Energy contribution | -12.62 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11293132 103 - 24543557 -----CCUGUCUAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC--UCACUCAAC-----GUCGACUUCCCGUUAGCACGCGGCU-AACUUG -----...(((..(((((....((((......((.(((((((......))))))).))....))))..--..)))))..-----...))).....((((((.....)))-)))... ( -27.80, z-score = -2.23, R) >droPer1.super_20 1505985 100 + 1872136 -----GCUGUCUAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC--UCACUCAAC-----GGCGACUUCCCGUUAGCACCUAGACCCA---- -----...(((((.((......((((......((.(((((((......))))))).))....))))((--(.....(((-----((.(....))))))))).)))))))...---- ( -30.00, z-score = -2.76, R) >dp4.chrXR_group6 2900253 100 + 13314419 -----GCUGUCUAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC--UCACUCAAC-----GGCGACUUCCCGUUAGCACCUAGACCCA---- -----...(((((.((......((((......((.(((((((......))))))).))....))))((--(.....(((-----((.(....))))))))).)))))))...---- ( -30.00, z-score = -2.76, R) >droAna3.scaffold_13337 5577601 105 - 23293914 -----CCUGUCCAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC--UCACUCAACGGCGAGGCGACUUCCCGUUAGCACCUAAGCCCC---- -----.........((((....((((......((.(((((((......))))))).))....))))((--(.....(((((.(((....))).))))))))......)))).---- ( -32.80, z-score = -2.32, R) >droEre2.scaffold_4784 11296912 102 - 25762168 ------CUGUCUAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC--UCACUCAAC-----GUCGACUUCCCGUUAGCACGCGGCU-AACUUG ------..(((..(((((....((((......((.(((((((......))))))).))....))))..--..)))))..-----...))).....((((((.....)))-)))... ( -27.80, z-score = -2.26, R) >droYak2.chr3L 11328009 103 - 24197627 -----CCUGUCUAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC--UCACUCAAC-----GUCGACUUCCCGUUAGCACGCGGCU-AACUUG -----...(((..(((((....((((......((.(((((((......))))))).))....))))..--..)))))..-----...))).....((((((.....)))-)))... ( -27.80, z-score = -2.23, R) >droSec1.super_0 3506202 104 - 21120651 -----CCUGUCUAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC--UCACUCAAC-----GUCGACUUCCCGUUAGCACGCGGCUAAACUUG -----...(((((.......))))).......((.(((((((......))))))).)).(((((((((--...((.(((-----(.........))))))...))))))))).... ( -27.90, z-score = -2.22, R) >droSim1.chr3L 10681664 104 - 22553184 -----CCUGUCUAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC--UCACUCAAC-----GUCGACUUCCCGUUAGCACGCGGCUAAACUUG -----...(((((.......))))).......((.(((((((......))))))).)).(((((((((--...((.(((-----(.........))))))...))))))))).... ( -27.90, z-score = -2.22, R) >droWil1.scaffold_180698 9744984 103 + 11422946 GACUCUCUGUCCAUGGAUAUUACG-UUUAUCAGAUGUGCAUUCAUUGAAAUGCACUUCCUUUCG-GAC--UCGCUCAAUGGCCAGGCCAUUUCCCGUUAGCACCUAC--------- ........((((...((((.....-..)))).((.(((((((......))))))).)).....)-)))--..(((.(((((............))))))))......--------- ( -23.10, z-score = -1.06, R) >droVir3.scaffold_13049 1582870 90 + 25233164 -----GCUGUCUAUGGGCAUUAUG-UUUAUCAGAUGUGCAUUCAUUCGAAUGCACUUCCUUUGGGCGCA----CUCAAC-----GGCGACUUCCCGUUAGAGCUG----------- -----(((((((((((((.....)-))))).))))((((((((....))))))))........)))((.----((.(((-----((.(....)))))))).))..----------- ( -29.00, z-score = -2.39, R) >droMoj3.scaffold_6680 6503479 104 + 24764193 -----GCUGUCUAUGGGUAUCAUG-UUUAUCAGAUGUGCAGCCAUUCGAAUGCACUUCCUUUGAGCGCAGUCCGUCGAG-----UGCGAUUUCCCGUUAGAGCGAAGCGAAGCAG- -----(((.((((((((.(((..(-((((...((.(((((..(....)..))))).))...)))))(((.((....)).-----))))))..)))).))))....))).......- ( -31.00, z-score = -0.86, R) >droGri2.scaffold_15110 14020347 93 - 24565398 ----------UUAUGGGUAUUAUA-UUUAUCAGAUGUGCAUUCAUUCGAAUGCACUUCCUUUGG-GGC--GCACUCAAC-----GGCGACUUCCCGUUAGCAGCCACAACUA---- ----------.....((((.....-..)))).((.((((((((....)))))))).))..(((.-(((--...((.(((-----((.(....))))))))..))).)))...---- ( -26.20, z-score = -2.32, R) >consensus _____CCUGUCUAUGGGUAUUAGGCUUUAUCAGAUGUGCAUUCAUUCAAAUGCACUUCCUUUGGCCGC__UCACUCAAC_____GGCGACUUCCCGUUAGCACGCAGCU_AA____ ........(((((((((...............((.(((((((......))))))).))..((((..........))))..............)))).))).))............. (-12.30 = -12.62 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:14 2011