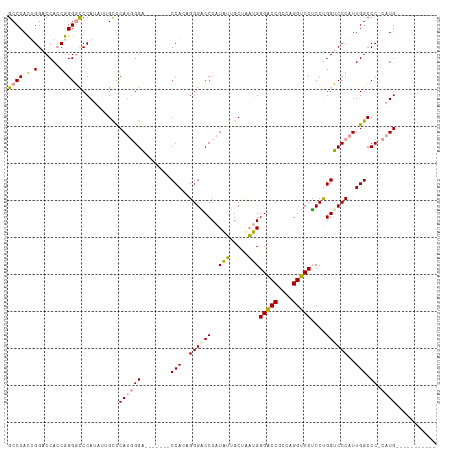
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,848,243 – 4,848,369 |
| Length | 126 |
| Max. P | 0.982448 |

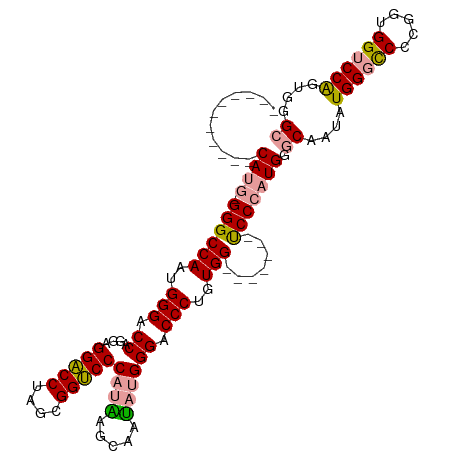
| Location | 4,848,243 – 4,848,340 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.26 |
| Shannon entropy | 0.44472 |
| G+C content | 0.64169 |
| Mean single sequence MFE | -53.93 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.71 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4848243 97 + 23011544 -----------CAUG-GGGCCAAUGGGACCAGGAGGACCUCGCGGUCCCAUAAGCAAUAUGGGUCCCUGUGG-------UCCCAUGAGCAGUAUGGGCCCCGGUGGUCCAGUGGGC -----------((((-((((((..((((((....(((((....)))))((((.....))))))))))..)))-------))))))).((....((((((.....))))))....)) ( -51.70, z-score = -2.92, R) >droSim1.chr2L 4718127 97 + 22036055 -----------CAUG-GGGCCAAUGGGACCAGGAGGACCUGGCGGUCCCAUGAGCAAUAUGGGCCCCGGUGG-------UCCCAUGGGCAAUAUGGGUCCCGGUGGUCCAGUGGGC -----------((((-(((((..((((.((....)).))))..))))))))).((....((((((((((.((-------.((((((.....)))))))))))).))))))....)) ( -52.50, z-score = -2.21, R) >droAna3.scaffold_12943 2587797 115 + 5039921 CUUCGGAGGACCAUUUGGGCCAAUGGGACCGGGUGGUCCAAAUGGACCCUGUGGCCCAAUGGGACCCUUUGGACCAGGUCCCUUUGGACCAGGUGGACCCUG-UGGACCGAUGGGC .((((.(((.((((((((.((((.((((((...(((((((((.((.(((...........))).)).))))))))))))))).)))).))))))))..))).-))))((....)). ( -57.60, z-score = -1.97, R) >consensus ___________CAUG_GGGCCAAUGGGACCAGGAGGACCUAGCGGUCCCAUAAGCAAUAUGGGACCCUGUGG_______UCCCAUGGGCAAUAUGGGCCCCGGUGGUCCAGUGGGC ..................(((.((.((((((.(.(((((....))))))...........((((((((((((.........)))))).......))))))...)))))).)).))) (-30.14 = -30.71 + 0.57)

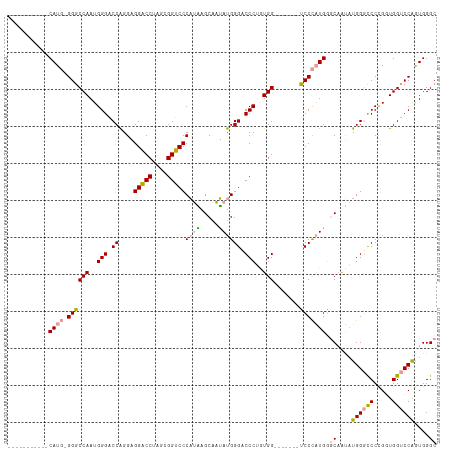
| Location | 4,848,243 – 4,848,340 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.26 |
| Shannon entropy | 0.44472 |
| G+C content | 0.64169 |
| Mean single sequence MFE | -47.53 |
| Consensus MFE | -29.14 |
| Energy contribution | -26.93 |
| Covariance contribution | -2.21 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4848243 97 - 23011544 GCCCACUGGACCACCGGGGCCCAUACUGCUCAUGGGA-------CCACAGGGACCCAUAUUGCUUAUGGGACCGCGAGGUCCUCCUGGUCCCAUUGGCCC-CAUG----------- ((((...((....)).))))..........((((((.-------(((..((((((((((.....))))(((((....)))))....))))))..))).))-))))----------- ( -46.50, z-score = -3.07, R) >droSim1.chr2L 4718127 97 - 22036055 GCCCACUGGACCACCGGGACCCAUAUUGCCCAUGGGA-------CCACCGGGGCCCAUAUUGCUCAUGGGACCGCCAGGUCCUCCUGGUCCCAUUGGCCC-CAUG----------- ((....(((.((.((((..(((((.......))))).-------...)))))).)))....)).((((((.((((((((....))))))......)).))-))))----------- ( -39.90, z-score = -0.76, R) >droAna3.scaffold_12943 2587797 115 - 5039921 GCCCAUCGGUCCA-CAGGGUCCACCUGGUCCAAAGGGACCUGGUCCAAAGGGUCCCAUUGGGCCACAGGGUCCAUUUGGACCACCCGGUCCCAUUGGCCCAAAUGGUCCUCCGAAG .....((((....-..(((.(((..(((.((((.(((((((((((((((.((.(((..((.....))))).)).)))))))))...)))))).)))).)))..))).))))))).. ( -56.20, z-score = -2.83, R) >consensus GCCCACUGGACCACCGGGGCCCAUAUUGCCCAUGGGA_______CCACAGGGACCCAUAUUGCUAAUGGGACCGCCAGGUCCUCCUGGUCCCAUUGGCCC_CAUG___________ .......((.(((..((((((.............((........))...((((....(((.....)))(((((....)))))))))))))))..)))))................. (-29.14 = -26.93 + -2.21)

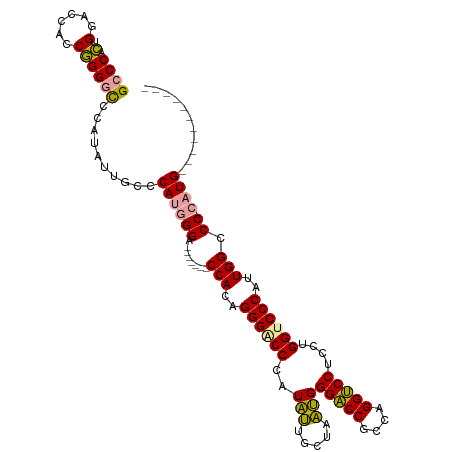
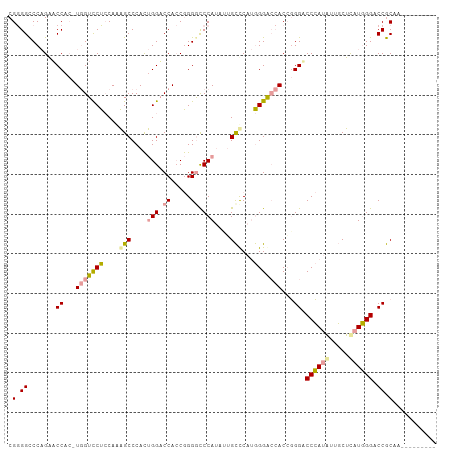
| Location | 4,848,270 – 4,848,369 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 64.13 |
| Shannon entropy | 0.48617 |
| G+C content | 0.63686 |
| Mean single sequence MFE | -43.41 |
| Consensus MFE | -22.46 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4848270 99 - 23011544 CGGGGCCCAGAACCACGUGGUCCUCCAAAGCCCACUGGACCACCGGGGCCCAUACUGCUCAUGGGACCACAGGGACCCAUAUUGCUUAUGGGACCGCGA--------- ((.(((((....((..(((((((.............))))))).))(((((((.......))))).))...))).((((((.....)))))).)).)).--------- ( -40.62, z-score = -1.24, R) >droSim1.chr2L 4718154 99 - 22036055 CGGGGCCCAGAUCCACCUGGUCCUCCAAAGCCCACUGGACCACCGGGACCCAUAUUGCCCAUGGGACCACCGGGGCCCAUAUUGCUCAUGGGACCGCCA--------- .((((((((((....((((((..((((........))))..)))))).........((((.(((.....))))))).........)).)))).)).)).--------- ( -37.40, z-score = 0.39, R) >droAna3.scaffold_12943 2587836 102 - 5039921 CGAGGUCCCGGACCA--UAGUUCU-CAGGGCCCAUCGGUCCA-CAGGGUCCACCUGGUCCAAAGGG--ACCUGGUCCAAAGGGUCCCAUUGGGCCACAGGGUCCAUUU ...((.((((((((.--..(....-).(((((....))))).-...)))))...((((((((.(((--((((........))))))).))))))))..))).)).... ( -52.20, z-score = -3.63, R) >consensus CGGGGCCCAGAACCAC_UGGUCCUCCAAAGCCCACUGGACCACCGGGGCCCAUAUUGCCCAUGGGACCACCGGGACCCAUAUUGCUCAUGGGACCGCAA_________ .(.((.......((...(((((((....(((....(((.((.....)).)))....)))...)))))))...)).((((((.....)))))).)).)........... (-22.46 = -23.03 + 0.57)

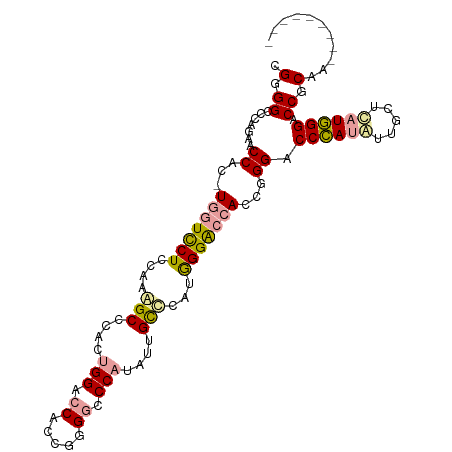
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:34 2011