| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,239,470 – 11,239,566 |
| Length | 96 |
| Max. P | 0.748104 |

| Location | 11,239,470 – 11,239,566 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.74 |
| Shannon entropy | 0.65389 |
| G+C content | 0.54125 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
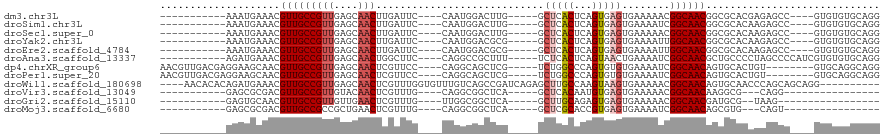
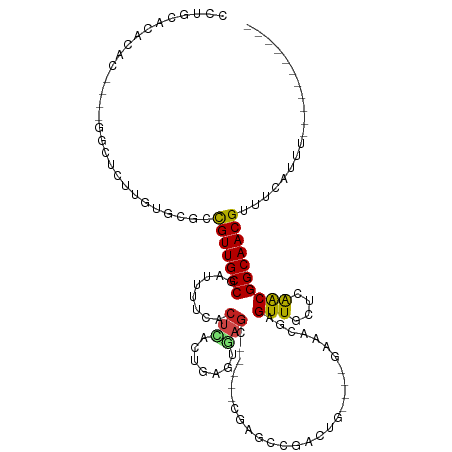
>dm3.chr3L 11239470 96 + 24543557 -----------AAAUGAAACGUUGCCGUUGAGCAACUUGAUUC----CAAUGGACUUG-----GCUCACUCAGUGAGUGAAAAACGGCAACGGCGCACGAGAGCC----GUGUGUGCAGG -----------........((((((((((............((----(...))).((.-----((((((...)))))).)).))))))))))(((((((.....)----))))))..... ( -34.60, z-score = -2.13, R) >droSim1.chr3L 10633772 96 + 22553184 -----------AAAUGAAACGUUGCCGUUGAGCAACUUGAUUC----CAAUGGACUUG-----GCUCACUCAGUGAGUGAAAAUCGGCAACGGCGCACAAGAGCC----GUGUGUGCAGG -----------........(((((((((..(((...(((....----)))..).))..-----))((((((...)))))).....)))))))(((((((......----.)))))))... ( -34.80, z-score = -2.28, R) >droSec1.super_0 3461189 96 + 21120651 -----------AAAUGAAACGUUGCCGUUGAGCAACUUGAUUC----CAAUGGACUUG-----GCUCACUCAGUGAGUGAAAAACGGCAACGGCGCACAAGAGCC----GUGUGUGCAGG -----------........((((((((((............((----(...))).((.-----((((((...)))))).)).))))))))))(((((((......----.)))))))... ( -34.80, z-score = -2.33, R) >droYak2.chr3L 11280166 96 + 24197627 -----------AAAUGAAACGUUGCCGUUGAGCAACUUGAUUC----CAAUGGACGCG-----GCUCACUCAGUGAGUGAAAAUUGGCAACGGCGCACAAGAGCC----GUGUGUGCAGG -----------........(((((((((((..(...(((....----)))..)...))-----))((((((...)))))).....)))))))(((((((......----.)))))))... ( -31.70, z-score = -0.97, R) >droEre2.scaffold_4784 11250414 96 + 25762168 -----------AAAUGAAACGUUGCCGUUGAGCAACUUGAUUC----CAAUGGACGCG-----GCUCACUCAGUGAGUGAAAAUUGGCAACGGCGCACAAGAGCC----GUGUGUGCAGG -----------........(((((((((((..(...(((....----)))..)...))-----))((((((...)))))).....)))))))(((((((......----.)))))))... ( -31.70, z-score = -0.97, R) >droAna3.scaffold_13337 5534574 100 + 23293914 -----------AGAUGAAACGUUGCCGUUGAGCAACUGGCUUC----CAGGCCGCUUU-----UCUCACUCAGUAACUGAAAAUCGGCAACGCUGCCCCUAGCCCCAUCGUGUGUGCAGG -----------.((((....(((((((.((((....(((((..----..)))))....-----.)))).((((...))))....)))))))((((....))))..))))........... ( -28.60, z-score = -0.49, R) >dp4.chrXR_group6 2854433 103 - 13314419 AACGUUGACGAGGAAGCAACGUUGCCGUUGAGCAACUCGUUCC----CAGGCAGCUCG-----UCUGGCCCAGUGUGUGAAAAUCGGCAACAGUGCACUGU--------GUGCAGGCAGG ...((((.(......)))))((((((...((((.....)))).----..))))))..(-----((((((.(((((..((.....)(....).)..))))).--------)).)))))... ( -34.80, z-score = 0.15, R) >droPer1.super_20 1458764 103 - 1872136 AACGUUGACGAGGAAGCAACGUUGCCGUUGAGCAACUCGUUCC----CAGGCAGCUCG-----UCUGGCCCAGUGUGUGAAAAUCGGCAACAGUGCACUGU--------GUGCAGGCAGG ...((((.(......)))))((((((...((((.....)))).----..))))))..(-----((((((.(((((..((.....)(....).)..))))).--------)).)))))... ( -34.80, z-score = 0.15, R) >droWil1.scaffold_180698 1635202 106 + 11422946 ----AACACACAGAUGAAACGUUGCCGUUGAGCAACUCGUUUGGUGUUUGUCAGCCGAUCAGAGCUUGCCAAGUAAGUGAAAAACGGCAACAGUGCAACCCAGCAGCAGG---------- ----(((((.(((((((...(((((......)))))))))))))))))(((..((((......((((((...))))))......)))).....(((......))))))..---------- ( -30.70, z-score = -0.70, R) >droVir3.scaffold_13049 1542571 81 - 25233164 -----------GAGCGCGACGUUGCCGUUGUACAACUCGUUUG----CAGGCGGCUCA-----GCUCACAAUGUGAGUGAAAAACGGCAACAAGGCG---CAGG---------------- -----------..((((...(((((((((.....((((..(((----..(((......-----)))..)))...))))....)))))))))...)))---)...---------------- ( -32.70, z-score = -2.28, R) >droGri2.scaffold_15110 13973502 81 + 24565398 -----------GAGUGCAACGUUGCCGUUGUUGAACUCGUUUG----UUGGCGGCUCA-----GCUUGCAGAGUGAGUGAAAAACGGCAACGAUGCG--UAAG----------------- -----------....(((.((((((((((.....(((((((((----(.(((......-----))).))))).)))))....)))))))))).))).--....----------------- ( -36.60, z-score = -4.23, R) >droMoj3.scaffold_6680 6454149 81 - 24764193 -----------GAGCGCGACGUUGCCGCCGCUGAACUCGUUUG----CAGGCGGCUCA-----GCUCGCACCGUGAGUGAAAAUCGGCAACAGCGUG---CAGU---------------- -----------..(((((..((((((((((((...(......)----..))))))(((-----.(((((...)))))))).....))))))..))))---)...---------------- ( -37.20, z-score = -1.73, R) >consensus ___________AAAUGAAACGUUGCCGUUGAGCAACUCGUUUC____CAGGCGGCUCG_____GCUCACUCAGUGAGUGAAAAUCGGCAACGGCGCACAAGAGCC____GUGUGUGCAGG ....................(((((((((....)))............................(((((...)))))........))))))............................. (-12.46 = -12.73 + 0.27)
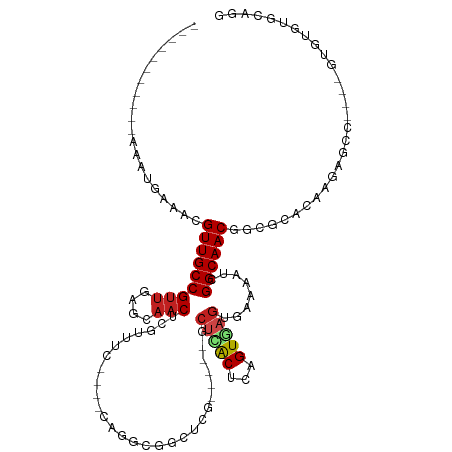
| Location | 11,239,470 – 11,239,566 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.74 |
| Shannon entropy | 0.65389 |
| G+C content | 0.54125 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -11.58 |
| Energy contribution | -11.53 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11239470 96 - 24543557 CCUGCACACAC----GGCUCUCGUGCGCCGUUGCCGUUUUUCACUCACUGAGUGAGC-----CAAGUCCAUUG----GAAUCAAGUUGCUCAACGGCAACGUUUCAUUU----------- ...((((....----.......))))..((((((((((.(((((((...)))))))(-----(((.....)))----).............))))))))))........----------- ( -29.70, z-score = -1.91, R) >droSim1.chr3L 10633772 96 - 22553184 CCUGCACACAC----GGCUCUUGUGCGCCGUUGCCGAUUUUCACUCACUGAGUGAGC-----CAAGUCCAUUG----GAAUCAAGUUGCUCAACGGCAACGUUUCAUUU----------- ...(((((...----......)))))..((((((((((((((((((...)))))))(-----(((.....)))----)))))..(((....))))))))))........----------- ( -29.70, z-score = -1.86, R) >droSec1.super_0 3461189 96 - 21120651 CCUGCACACAC----GGCUCUUGUGCGCCGUUGCCGUUUUUCACUCACUGAGUGAGC-----CAAGUCCAUUG----GAAUCAAGUUGCUCAACGGCAACGUUUCAUUU----------- ...(((((...----......)))))..((((((((((.(((((((...)))))))(-----(((.....)))----).............))))))))))........----------- ( -30.70, z-score = -2.17, R) >droYak2.chr3L 11280166 96 - 24197627 CCUGCACACAC----GGCUCUUGUGCGCCGUUGCCAAUUUUCACUCACUGAGUGAGC-----CGCGUCCAUUG----GAAUCAAGUUGCUCAACGGCAACGUUUCAUUU----------- ...(((((...----......)))))..(((((((.......((((...))))((((-----.((.(((...)----)).....)).))))...)))))))........----------- ( -27.90, z-score = -1.13, R) >droEre2.scaffold_4784 11250414 96 - 25762168 CCUGCACACAC----GGCUCUUGUGCGCCGUUGCCAAUUUUCACUCACUGAGUGAGC-----CGCGUCCAUUG----GAAUCAAGUUGCUCAACGGCAACGUUUCAUUU----------- ...(((((...----......)))))..(((((((.......((((...))))((((-----.((.(((...)----)).....)).))))...)))))))........----------- ( -27.90, z-score = -1.13, R) >droAna3.scaffold_13337 5534574 100 - 23293914 CCUGCACACACGAUGGGGCUAGGGGCAGCGUUGCCGAUUUUCAGUUACUGAGUGAGA-----AAAGCGGCCUG----GAAGCCAGUUGCUCAACGGCAACGUUUCAUCU----------- ...........(((((.((.....))((((((((((...(((((...)))))((((.-----..((((((...----...))).))).)))).))))))))))))))).----------- ( -34.20, z-score = -0.85, R) >dp4.chrXR_group6 2854433 103 + 13314419 CCUGCCUGCAC--------ACAGUGCACUGUUGCCGAUUUUCACACACUGGGCCAGA-----CGAGCUGCCUG----GGAACGAGUUGCUCAACGGCAACGUUGCUUCCUCGUCAACGUU ..((((((...--------.))).)))..(((((((...........(((...))).-----.((((.((.((----....)).)).))))..)))))))(((((......).))))... ( -30.60, z-score = 0.32, R) >droPer1.super_20 1458764 103 + 1872136 CCUGCCUGCAC--------ACAGUGCACUGUUGCCGAUUUUCACACACUGGGCCAGA-----CGAGCUGCCUG----GGAACGAGUUGCUCAACGGCAACGUUGCUUCCUCGUCAACGUU ..((((((...--------.))).)))..(((((((...........(((...))).-----.((((.((.((----....)).)).))))..)))))))(((((......).))))... ( -30.60, z-score = 0.32, R) >droWil1.scaffold_180698 1635202 106 - 11422946 ----------CCUGCUGCUGGGUUGCACUGUUGCCGUUUUUCACUUACUUGGCAAGCUCUGAUCGGCUGACAAACACCAAACGAGUUGCUCAACGGCAACGUUUCAUCUGUGUGUU---- ----------(((......)))..((((((((((((((....((((..((((..((((......))))........))))..)))).....))))))))))........))))...---- ( -24.30, z-score = 1.34, R) >droVir3.scaffold_13049 1542571 81 + 25233164 ----------------CCUG---CGCCUUGUUGCCGUUUUUCACUCACAUUGUGAGC-----UGAGCCGCCUG----CAAACGAGUUGUACAACGGCAACGUCGCGCUC----------- ----------------...(---(((..((((((((((...(((((...((((..((-----......))..)----)))..))).))...))))))))))..))))..----------- ( -32.60, z-score = -3.42, R) >droGri2.scaffold_15110 13973502 81 - 24565398 -----------------CUUA--CGCAUCGUUGCCGUUUUUCACUCACUCUGCAAGC-----UGAGCCGCCAA----CAAACGAGUUCAACAACGGCAACGUUGCACUC----------- -----------------....--.(((.((((((((((.((.((((.(((.......-----.)))..(....----)....))))..)).)))))))))).)))....----------- ( -25.50, z-score = -3.43, R) >droMoj3.scaffold_6680 6454149 81 + 24764193 ----------------ACUG---CACGCUGUUGCCGAUUUUCACUCACGGUGCGAGC-----UGAGCCGCCUG----CAAACGAGUUCAGCGGCGGCAACGUCGCGCUC----------- ----------------...(---(.((.((((((((..((.(((.....))).))((-----(((((((....----....)).)))))))..)))))))).)).))..----------- ( -32.30, z-score = -0.72, R) >consensus CCUGCACACAC____GGCUCUUGUGCGCCGUUGCCGAUUUUCACUCACUGAGUGAGC_____CGAGCCGACUG____GAAACGAGUUGCUCAACGGCAACGUUUCAUUU___________ ............................(((((((........(((.......)))............................(((....))))))))))................... (-11.58 = -11.53 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:04 2011