| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,230,317 – 11,230,414 |
| Length | 97 |
| Max. P | 0.997603 |

| Location | 11,230,317 – 11,230,414 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.92 |
| Shannon entropy | 0.22273 |
| G+C content | 0.33630 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -17.31 |
| Energy contribution | -17.59 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.997603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
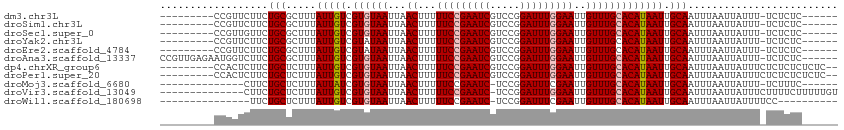
>dm3.chr3L 11230317 97 + 24543557 ---------CCGUUCUUCUGCGCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUU-UCUCUC------ ---------............((...(((((.((((((...((...(((((((((.....)))))))))..))))))))))))).)).............-......------ ( -20.90, z-score = -3.32, R) >droSim1.chr3L 10624283 97 + 22553184 ---------CCGUUCUUCUGCGCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUU-UCUCUC------ ---------............((...(((((.((((((...((...(((((((((.....)))))))))..))))))))))))).)).............-......------ ( -20.90, z-score = -3.32, R) >droSec1.super_0 3452059 97 + 21120651 ---------CCGUUGUUCUGCGCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUU-UCUCUC------ ---------............((...(((((.((((((...((...(((((((((.....)))))))))..))))))))))))).)).............-......------ ( -20.90, z-score = -2.91, R) >droYak2.chr3L 11270193 97 + 24197627 ---------CCGUUCUUCUGCGCUUUAUUGUCGUAUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUU-UCUCUC------ ---------.........(((((......).))))(((((((....(((((((((.....)))))))))....(((((......))))).)))))))...-......------ ( -18.30, z-score = -2.51, R) >droEre2.scaffold_4784 11241227 97 + 25762168 ---------CCGUUCUUCUGCGCUUUAUUGUCGUAUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUU-UCUCUC------ ---------.........(((((......).))))(((((((....(((((((((.....)))))))))....(((((......))))).)))))))...-......------ ( -18.30, z-score = -2.51, R) >droAna3.scaffold_13337 5523951 106 + 23293914 CCGUUGAGAAUGGUCUUCUGCGCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUU-UCUCUC------ .....(((((((((....((((....(((((.((((((...((...(((((((((.....)))))))))..))))))))))))))))).....)))))))-))....------ ( -25.90, z-score = -3.09, R) >dp4.chrXR_group6 2845394 102 - 13314419 ---------CCACUCUUCUGCUCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUUCUCUCUCUCUC-- ---------.........(((.....(((((.((((((...((...(((((((((.....)))))))))..))))))))))))).))).......................-- ( -20.10, z-score = -4.25, R) >droPer1.super_20 1449666 102 - 1872136 ---------CCACUCUUCUGCUCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUUCUCUCUCUCUC-- ---------.........(((.....(((((.((((((...((...(((((((((.....)))))))))..))))))))))))).))).......................-- ( -20.10, z-score = -4.25, R) >droMoj3.scaffold_6680 6444982 91 - 24764193 --------------CUUCUGCUCUUUAUUAUCGUGUAAUUAACUUUUUCCGAAUC-UCCGGAUUUCGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUU-UCUUUC------ --------------....(((.....(((((.((((((...((...(((.(((((-....))))).)))..))))))))))))).)))............-......------ ( -14.50, z-score = -2.89, R) >droVir3.scaffold_13049 1534981 98 - 25233164 --------------CUUCUGCUCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUC-UCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUUCUUUUCUUUUUGU --------------....(((.....(((((.((((((...((...(((((((((-....)))))))))..))))))))))))).)))......................... ( -20.30, z-score = -4.42, R) >droWil1.scaffold_180698 1624125 87 + 11422946 ---------------UUCUGCUCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUC-UCCGGAUUUCGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUUUCC---------- ---------------...(((.....(((((.((((((...((...(((.(((((-....))))).)))..))))))))))))).)))...............---------- ( -14.50, z-score = -2.41, R) >consensus _________CCGUUCUUCUGCGCUUUAUUGUCGUGUAAUUAACUUUUUCCGAAUCGUCCGGAUUUGGAAUUGUUUGCACAUAAUUGCAAUUUAAUUAUUU_UCUCUC______ ..................(((.....(((((.((((((...((...(((((((((.....)))))))))..))))))))))))).)))......................... (-17.31 = -17.59 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:02 2011