
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,212,596 – 11,212,771 |
| Length | 175 |
| Max. P | 0.854038 |

| Location | 11,212,596 – 11,212,703 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 68.26 |
| Shannon entropy | 0.61091 |
| G+C content | 0.42596 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -10.01 |
| Energy contribution | -8.85 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.63 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11212596 107 - 24543557 GGCGUGCCCAAAUCAUAAUCAAAUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUG--GCCGCUGGCCAAGAAUGAA---UGCCUUAAAUUUU ...((.(((((............))))).))(((((.....((((.-(((((((...))))))).))))(((((((--(((...))))))))))...---.)))))....... ( -36.20, z-score = -3.51, R) >droSim1.chr3L 10606901 107 - 22553184 GGCGUGCCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUG--GCCGCUGGCCAACAAUUAA---UGCCACAAAUUUU (((((.(((((((........)))))))..((.((((((((((((.-(((((((...))))))).)))).)).)))--)))..))...........)---))))......... ( -34.00, z-score = -3.03, R) >droSec1.super_0 3434698 107 - 21120651 GGCGUGUCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUUUUG--GCCGCUGGCCAACAAUUAA---UGCCACAAAUUUU (((((..((((((........))))))..))..((((((((((((.-(((((((...))))))).))))..)))))--))))))(((..........---.)))......... ( -33.60, z-score = -3.00, R) >droYak2.chr3L 11251429 107 - 24197627 GGCGUGCCCAAAUCACGAUCAAUUUGGGAACAAGGCCAAAAGCAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUG--GACGCUGGCCAAGAAUUAA---UGCCACACAUUUU (((.(.(((((((........)))))))....).)))....(((..-(((((((...)))))))..((((((((((--(.(....))))))))))))---))).......... ( -32.30, z-score = -2.19, R) >droEre2.scaffold_4784 11222731 107 - 25762168 GGCAUGCCCAAAUCAUUAUCAAUUUGGGAACAAGGCCAAAACCAAG-AGAUUUGUGCCGAAUCUGUUUGAUUCUUG--GGCGCUGGCCAAGAAUUAA---UGCCACACAUUUU (((((.(((((((........))))))).....(((((....(...-.)....((((((((((.....)))))...--))))))))))........)---))))......... ( -33.90, z-score = -2.37, R) >droAna3.scaffold_13337 5506489 110 - 23293914 GGCAUACCGAAACU-UAAUCAAUUUUAGAGCAAGGCCAUAACCAAGAAGAUUUGUUCUGAUUCCAUUUGAUUCUUG--GCUAUACGCCAAGAAAUAAUUUUUCUCUAAGAUUA ((....))......-.....(((((((((((((((......)).((((......))))........))).((((((--((.....)))))))).........)))))))))). ( -22.90, z-score = -1.93, R) >dp4.chrXR_group6 2824268 98 + 13314419 AGCAGGCCUAAACA-CGAUCAAUUUGGGAACAAGGCCACAGCGGAGGGCCCCCCUUUGGAGGCUUGCCAAGAAUUAAUGUUUCACAUUUAUAAAGA----UUC---------- .((((((((.....-.......((((....)))).......(((((((...))))))).))))))))...(((((((((.....))))......))----)))---------- ( -25.10, z-score = -0.44, R) >droPer1.super_20 1428797 98 + 1872136 AGCAGGCCUAAACA-CGAUCAAUUUGGGAACAAGGCCACGGCGGAGGGCCCCCCUUUGGAGGCUUGCCAAGAAUUAAUGUUCCACAUUUAUAAAGA----UUC---------- .((((((((.....-((.....((((....))))....)).(((((((...))))))).))))))))...((((....))))..............----...---------- ( -26.20, z-score = -0.20, R) >consensus GGCAUGCCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG_AGAUUUGUUCCGAAUCUGUUUGAUUCUUG__GCCGCUGGCCAAGAAUUAA___UGCCACAAAUUUU (((...((((((..........))))))...............(((.(((((((...))))))).))).................)))......................... (-10.01 = -8.85 + -1.16)


| Location | 11,212,624 – 11,212,743 |
|---|---|
| Length | 119 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.60499 |
| G+C content | 0.39870 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -9.25 |
| Energy contribution | -8.56 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.569289 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 11212624 119 - 24543557 CAAAAUCAAGAACCUACAAAUUUAAUAAUACAACUAAUUGGGCGUGCCCAAAUCAUAAUCAAAUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUGGCCG .....................................(((((....)))))............(((....)))((((((((((((.-(((((((...))))))).)))).)).)))))). ( -27.70, z-score = -2.28, R) >droSim1.chr3L 10606929 113 - 22553184 ------CAAGAGCCUACAAAUUGAAUAAUACAACUAAUUGGGCGUGCCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUGGCCG ------.....(((((....(((.......))).....)))))((.(((((((........))))))).))..((((((((((((.-(((((((...))))))).)))).)).)))))). ( -33.70, z-score = -3.63, R) >droSec1.super_0 3434726 113 - 21120651 ------CAAGAGCCUACAAAUUGAAUAAUACAACUAAUUGGGCGUGUCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUUUUGGCCG ------.....(((((....(((.......))).....)))))((..((((((........))))))..))..((((((((((((.-(((((((...))))))).))))..)))))))). ( -33.50, z-score = -3.68, R) >droYak2.chr3L 11251457 119 - 24197627 CAUAUUCAAGAGCCAACAAAUUUAAUAAUACAACUAAUUAGGCGUGCCCAAAUCACGAUCAAUUUGGGAACAAGGCCAAAAGCAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUGGACG ....(((((((((.(((.......................(((.(.(((((((........)))))))....).))).........-(((((((...)))))))))).).)))))))).. ( -27.10, z-score = -2.11, R) >droEre2.scaffold_4784 11222759 119 - 25762168 CAUAUUCAAGAGCCCACAAAUUUAAUAAUACAACUAAUUAGGCAUGCCCAAAUCAUUAUCAAUUUGGGAACAAGGCCAAAACCAAG-AGAUUUGUGCCGAAUCUGUUUGAUUCUUGGGCG ...........((((((((((((..((((.......))))(((...(((((((........)))))))......))).........-))))))))...(((((.....)))))..)))). ( -30.90, z-score = -2.78, R) >droAna3.scaffold_13337 5506520 105 - 23293914 -------------UAAUGAAAU-AAUAAUAAAUCUAAUUAGGCAUACCGAAA-CUUAAUCAAUUUUAGAGCAAGGCCAUAACCAAGAAGAUUUGUUCUGAUUCCAUUUGAUUCUUGGCUA -------------..(((...(-(((..........))))..))).((((..-...((((((..((((((((((................))))))))))......)))))).))))... ( -12.19, z-score = 1.05, R) >dp4.chrXR_group6 2824305 96 + 13314419 ---UGACAAGAAACACCCAAAU-UUUGAU-CAACCAAUUAAGCAGGCCUAAA-CACGAUCAAUUUGGGAACAAGGCCACAGCGGAGGGCCCCCCUUUGGAGG------------------ ---((((((((..........)-)))).)-)).((.........(((((...-..(((.....)))......)))))....(((((((...)))))))..))------------------ ( -20.20, z-score = 0.37, R) >droPer1.super_20 1428834 96 + 1872136 ---UGACAAGAAACACCCAAAU-UUUGAU-CAACCAAUUAAGCAGGCCUAAA-CACGAUCAAUUUGGGAACAAGGCCACGGCGGAGGGCCCCCCUUUGGAGG------------------ ---((((((((..........)-)))).)-)).((......((.(((((...-..(((.....)))......)))))...))((((((...))))))))...------------------ ( -20.50, z-score = 0.69, R) >consensus ______CAAGAGCCAACAAAUU_AAUAAUACAACUAAUUAGGCAUGCCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG_AGAUUUGUUCCGAAUCUGUUUGAUUCUUGGCCG ........................................(((...((((((..........))))))......)))...........((((((...))))))................. ( -9.25 = -8.56 + -0.69)

| Location | 11,212,663 – 11,212,771 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.33 |
| Shannon entropy | 0.69537 |
| G+C content | 0.36350 |
| Mean single sequence MFE | -17.26 |
| Consensus MFE | -6.92 |
| Energy contribution | -6.94 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11212663 108 - 24543557 GAAAUACUUUAGAGACAGGUCUUGACGGCAAAAUCAAGAACCUACAAAUUUAAUAAUA--CAACUAAUUGGGCGUGCCCAAAUCAUAAUCAAAUUGGGAACAAGGCCAAA ........(((((...(((((((((........))))).)))).....))))).....--.......((((.(...(((((............))))).....).)))). ( -20.10, z-score = -1.43, R) >droSim1.chr3L 10606968 97 - 22553184 GAAAUAGUUUAGAGACAGGUCU-----------UCAAGAGCCUACAAAUUGAAUAAUA--CAACUAAUUGGGCGUGCCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAA ......(((....))).(((((-----------(.....(((((....(((.......--))).....)))))((.(((((((........))))))).))))))))... ( -22.80, z-score = -2.29, R) >droSec1.super_0 3434765 86 - 21120651 -----------GAAAUAGGUCU-----------ACAAGAGCCUACAAAUUGAAUAAUA--CAACUAAUUGGGCGUGUCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAA -----------......(((((-----------......(((((....(((.......--))).....)))))((..((((((........))))))..)).)))))... ( -19.80, z-score = -2.54, R) >droYak2.chr3L 11251496 108 - 24197627 GAAAUAUUUUAGAGACAGGUCGUGACGGCAUAUUCAAGAGCCAACAAAUUUAAUAAUA--CAACUAAUUAGGCGUGCCCAAAUCACGAUCAAUUUGGGAACAAGGCCAAA .................((((((((.(((((........(((................--..........))))))))....))))))))..(((((........))))) ( -20.77, z-score = -1.41, R) >droEre2.scaffold_4784 11222798 108 - 25762168 GAAAUAGUUUAGAGACAGGUCUUGACGGCAUAUUCAAGAGCCCACAAAUUUAAUAAUA--CAACUAAUUAGGCAUGCCCAAAUCAUUAUCAAUUUGGGAACAAGGCCAAA ......(((....))).(((((((..(((..........)))................--................(((((((........)))))))..)))))))... ( -22.00, z-score = -1.91, R) >droAna3.scaffold_13337 5506560 88 - 23293914 -------------GACAUACUUUCA------AGACAGUUUUUUAAUGAAAUAAUAAUA--AAUCUAAUUAGGCAUACCGAAA-CUUAAUCAAUUUUAGAGCAAGGCCAUA -------------............------.((.((((((...(((...((((....--......))))..)))...))))-))...)).................... ( -3.90, z-score = 1.74, R) >dp4.chrXR_group6 2824327 108 + 13314419 GAAAUA-UUUCUGCAAAUUUCCCAAGACAUUCCCCUGACAAGAAACACCCAAAUUUUGAUCAACCAAUUAAGCAGGCCUAAA-CACGAUCAAUUUGGGAACAAGGCCACA ......-.....((....((((((((........((....)).............((((((.....................-...))))))))))))))....)).... ( -14.36, z-score = 0.15, R) >droPer1.super_20 1428856 109 + 1872136 GAAAUAUUUUCUGCAAAUUUCCCAAGACAUUCCCCUGACAAGAAACACCCAAAUUUUGAUCAACCAAUUAAGCAGGCCUAAA-CACGAUCAAUUUGGGAACAAGGCCACG ............((....((((((((........((....)).............((((((.....................-...))))))))))))))....)).... ( -14.36, z-score = 0.11, R) >consensus GAAAUA_UUUAGAGACAGGUCUUGA_____UA_UCAAGAGCCUACAAAUUUAAUAAUA__CAACUAAUUAGGCAUGCCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAA ......................................................................(((...((((((..........))))))......)))... ( -6.92 = -6.94 + 0.02)

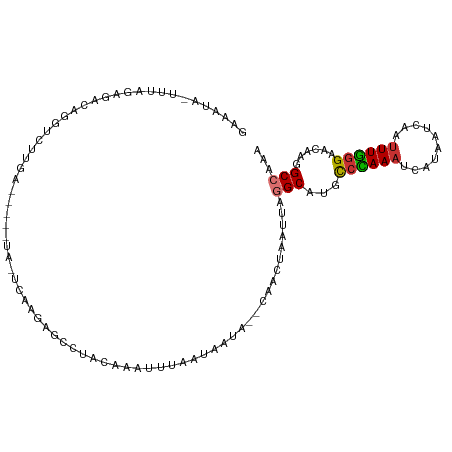

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:01 2011