| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,198,370 – 11,198,483 |
| Length | 113 |
| Max. P | 0.768676 |

| Location | 11,198,370 – 11,198,483 |
|---|---|
| Length | 113 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.99 |
| Shannon entropy | 0.72585 |
| G+C content | 0.41422 |
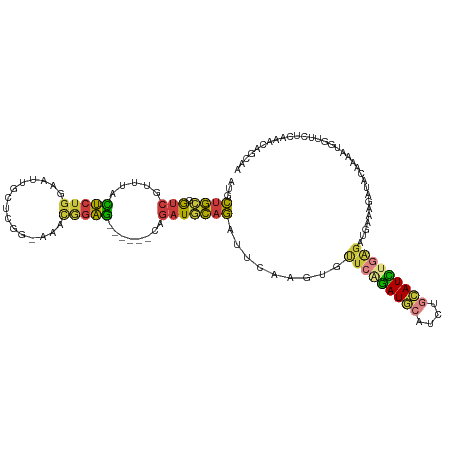
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -11.79 |
| Energy contribution | -11.82 |
| Covariance contribution | 0.03 |
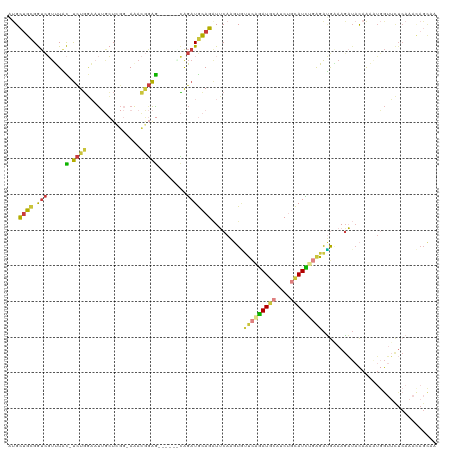
| Combinations/Pair | 1.67 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
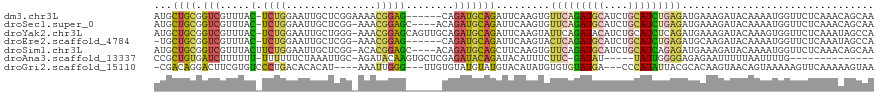
>dm3.chr3L 11198370 113 - 24543557 AUGCUGCGGUCGUUUAC-UCUGGAAUUGCUCGGAAAACGGAG------CAGAUGCAGAUUCAAGUGUUCAGAUGCAUCUGCAUCUGAGAUGAAAGAUACAAAAUGGUUCUCAAACAGCAA .(((((.(..(((((..-((((.(.((((((.(....).)))------))).).))))((((....(((((((((....))))))))).)))).......)))))..)......))))). ( -35.90, z-score = -2.14, R) >droSec1.super_0 3420651 114 - 21120651 AUGCUGCGGUCGUUUAC-UCUGGAAUUGCUCGG-AAACGGAGC----ACAGAUGCAGAUUCAAGUGUUCAGAUGCAUCUGCAUCUGAGAUGAAAGAUACAAAAUGGUUCUCAAACAGCAA .(((((...((((((..-........((((((.-...).))))----)(((((((((((.((..........)).))))))))))))))))).(((..(.....)..)))....))))). ( -35.90, z-score = -1.95, R) >droYak2.chr3L 11236791 118 - 24197627 AUGCUGCGGUCGUUUAC-UCUGGAAUUGCUGGG-AAACGGAGCAGUUGCAGAUGCAGAUUCAAGUAUUCAGAUACAUCUGCAUCUCAGAUGAAAGAUACAAAGUGGUUCUCAAAUAGCCA ..(((...(((.((((.-((((.(((((((.(.-...).).))))))..((((((((((....((((....)))))))))))))))))))))).)))....)))((((.......)))). ( -34.10, z-score = -1.17, R) >droEre2.scaffold_4784 11208101 111 - 25762168 -UGCUGCGGUCGUUUAC-UCUGGAAUUGCUCGG-AAACGGAG------CAGAUGCAGAUUCAAGUACUCAGAUGCAUCUGCAUCUGAGAUGCAAGAUACAAAAUGGUUCUCAAAUAGCCA -......(((..(((..-((((.(.(((((((.-...).)))------))).).)))).....((((((((((((....))))))))).)))...................)))..))). ( -38.40, z-score = -2.82, R) >droSim1.chr3L 10592866 115 - 22553184 AUGCUGCGGUCGUUUACUUCUGGAAUUGCUCGG-ACACGGAGC----ACAGAUGCAGCUUCAAGUGUUCAGAUGCAUCUGCAUCAGAGAUGAAAGAUACAAAAUGGUUCUCAAACAGCAA .(((((.(..(((((...(((.......(((((-(((((((((----.........)))))..)))))).(((((....))))).))).....)))....)))))..)......))))). ( -31.30, z-score = -0.21, R) >droAna3.scaffold_13337 5492087 98 - 23293914 CCGCUGUGAUCUUUUUU-UUUUUUCUAAAUUGC-AGAUACAAGUGCUCGAGAUACAGAUACAUUUCUUC-GAUAU-----UAUUGGGGAGAGAAUUUUUAAUUUUG-------------- .((((...((((.(..(-((......)))..).-))))...))))..((((((..((((...(((((((-(((..-----.))))))))))..))))...))))))-------------- ( -14.30, z-score = 0.61, R) >droGri2.scaffold_15110 10185384 109 + 24565398 -CGACAGGACUUCGUGUCCCUGACACACAU----AAAUUGGG---UUGUGUAUGUAUGUACAUAUGUGUGUAUGA---CCCAUAUUACGCACAAGUAACAGUAAAAAGUUCAAAAAGUAA -.....((((((.(((((...)))))((..----....((((---(..((((..((((....))))..))))..)---))))..((((......))))..))...))))))......... ( -26.20, z-score = -0.68, R) >consensus AUGCUGCGGUCGUUUAC_UCUGGAAUUGCUCGG_AAACGGAG______CAGAUGCAGAUUCAAGUGUUCAGAUGCAUCUGCAUCUGAGAUGAAAGAUACAAAAUGGUUCUCAAACAGCAA ...(((((..........((((...............))))...........))))).........(((((((((....)))))))))................................ (-11.79 = -11.82 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:57 2011