| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,189,579 – 11,189,658 |
| Length | 79 |
| Max. P | 0.703841 |

| Location | 11,189,579 – 11,189,658 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 52.38 |
| Shannon entropy | 0.65032 |
| G+C content | 0.41341 |
| Mean single sequence MFE | -12.53 |
| Consensus MFE | -3.95 |
| Energy contribution | -3.07 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
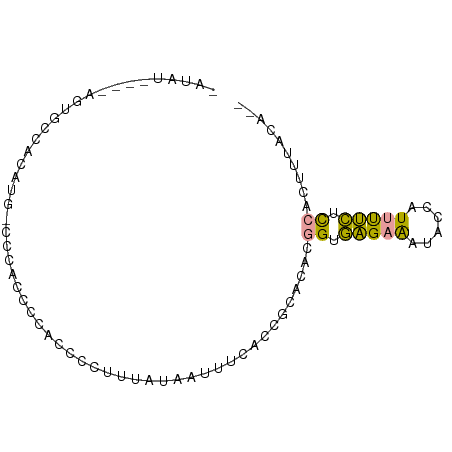
>dm3.chr3L 11189579 79 - 24543557 GAUAU----AGUGCCACAUG-CCCACCCCACCCCUUUAUAAUUUCACCGCACACGGUGAGAAAUACCAUUUUCUCCACUUUGCA-- (....----((((.......-......................((((((....))))))((((......))))..))))...).-- ( -12.30, z-score = -2.58, R) >droSim1.chr3L 10584144 84 - 22553184 CAUAUAGUGAGUGCCACAUGCCCCACCCCACCCCUUUAUAAUUUCGCGGCACACGGCGAGAAAUACCAUUUUCUCCACUUUACA-- ..((.((((...(((...((((.(.....................).))))...)))((((((......)))))))))).))..-- ( -17.00, z-score = -3.04, R) >apiMel3.Group10 3207710 74 - 11440700 ------------GAUAUAAUUCCAACAAUAUAGCUUCGUAUUAUAAAUGAUUAUAGUAGGAGAAACUGUCCCUUUCAAGUAUAGUA ------------.....................((((.((((((((....))))))))))))..((((((........).))))). ( -8.30, z-score = 0.18, R) >consensus _AUAU____AGUGCCACAUG_CCCACCCCACCCCUUUAUAAUUUCACCGCACACGGUGAGAAAUACCAUUUUCUCCACUUUACA__ ......................................................((.(((((......))))).)).......... ( -3.95 = -3.07 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:56 2011