| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,180,391 – 11,180,494 |
| Length | 103 |
| Max. P | 0.841599 |

| Location | 11,180,391 – 11,180,494 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Shannon entropy | 0.43322 |
| G+C content | 0.65354 |
| Mean single sequence MFE | -48.16 |
| Consensus MFE | -24.05 |
| Energy contribution | -24.29 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
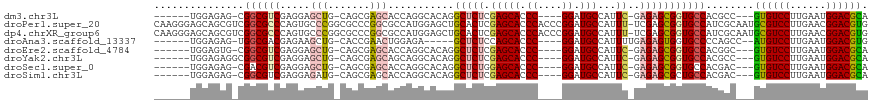
>dm3.chr3L 11180391 103 + 24543557 ------UGGAGAG-CGGCGUCGAGGAGCUG-CAGCGAGCACCAGGCACAGGCUCUCGAGCACCC----GGAUGCCAUUC-GAGAGCGGUGCCACGCC---GUGUCCUUGAAUGGACGCA ------.......-.(((((...((.(((.-.....))).)).(((((..(((((((((((...----...)))...))-)))))).))))))))))---((((((......)))))). ( -50.40, z-score = -2.67, R) >droPer1.super_20 1393210 118 - 1872136 CAAGGGAGCAGCGUCGGCGCCCAGUGCCCGGCGCCCGGCGCCAUGGAGCUGCACUCGAGCACCCACCCGGAUGCCAUUU-UCGAGCGGUGCCAUCGCAAUGCGUCCUUGAACGGACGUG (((((..((((((..((((((((((.((.(((((...)))))..)).))))..((((((((.((....)).))).....-))))).))))))..)))..)))..))))).......... ( -52.70, z-score = -1.16, R) >dp4.chrXR_group6 2789009 118 - 13314419 CAAGGGAGCAGCGUCGGCGCCCAGUGCCCGGCGCCCGGCGCCAUGGAGCUGCACUCGAGCACCCACCCGGAUGCCAUUU-UCGAGCGGUGCCAUCGCAAUGCGUCCUUGAACGGACGUG (((((..((((((..((((((((((.((.(((((...)))))..)).))))..((((((((.((....)).))).....-))))).))))))..)))..)))..))))).......... ( -52.70, z-score = -1.16, R) >droAna3.scaffold_13337 5473799 100 + 23293914 ------UGGAGAG-UGGCGACGAGAAGCUG-CACCGAACUGGAGA-----GCUCUCCAGCACCC----GGAUGCCAUUUUGAGAGUGGUGCCCCAGCC--AUGUCCUUGAAUGGACGUG ------.((...(-..((........))..-)......((((((.-----...))))))...))----((..(((((((...)))))))..))....(--((((((......))))))) ( -36.30, z-score = -1.94, R) >droEre2.scaffold_4784 11190013 103 + 25762168 ------UGGAGUG-CGGCGUCGAGGAGCUG-CAGCGAGCACCAGGCACAGGCUCUCGAGCACCC----GGAUGCCAUUC-GAGAGCGGUGCCACGGC---GUGUCCUUGAAUGGACGCA ------.....((-(.((((((.((.(((.-.....))).)).(((((..(((((((((((...----...)))...))-)))))).))))).))))---))((((......))))))) ( -49.10, z-score = -1.95, R) >droYak2.chr3L 11218017 104 + 24197627 ------UGGAGAGGCGGCGUCGAGGAGCUG-CAGCGAGCAGCAGGCACAGGCUCUCGAGCACCC----GGAUGCCAUUC-GAGAGCGGUGCCACGCC---GUGUCCUUGAAUGGACGCA ------.........(((((......((((-(.....))))).(((((..(((((((((((...----...)))...))-)))))).))))))))))---((((((......)))))). ( -53.60, z-score = -3.14, R) >droSec1.super_0 3403207 103 + 21120651 ------UGGAGAG-CGACGUCGAGGAGCUG-CAGCGAGCACCAGGCACAGGCUCUGGAGCACCC----GGAUGCCAUUC-GAGAGCGGUGCCACGAC---GUGUCCUUGAAUGGACGCA ------......(-(.((((((.((.(((.-.....))).)).(((((..(((((.(((((...----...)))...))-.))))).))))).))))---))((((......)))))). ( -45.70, z-score = -2.26, R) >droSim1.chr3L 10576268 103 + 22553184 ------UGGAGAG-CGGCGUCGAGGAGAUG-CAGCGAGCACCAGGCACAGGCUCUCGAGCACCC----GGAUGCCAUUC-GAGAGCGCUGCCACGAC---GUGUCCUUGAAUGGACGCA ------(((.(.(-(.(((((.....))))-).))...).)))((((...(((((((((((...----...)))...))-))))))..)))).....---((((((......)))))). ( -44.80, z-score = -1.93, R) >consensus ______UGGAGAG_CGGCGUCGAGGAGCUG_CAGCGAGCACCAGGCACAGGCUCUCGAGCACCC____GGAUGCCAUUC_GAGAGCGGUGCCACGGC___GUGUCCUUGAAUGGACGCA ...............((((((.....(((.......)))...........(((((...(((((.....)).))).......)))))))))))........((((((......)))))). (-24.05 = -24.29 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:54 2011