| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,173,001 – 11,173,104 |
| Length | 103 |
| Max. P | 0.666224 |

| Location | 11,173,001 – 11,173,104 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.80 |
| Shannon entropy | 0.28020 |
| G+C content | 0.47425 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -23.23 |
| Energy contribution | -23.26 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
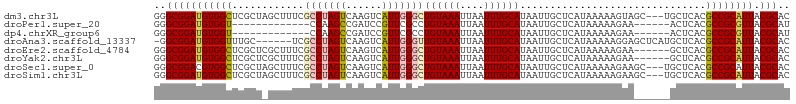
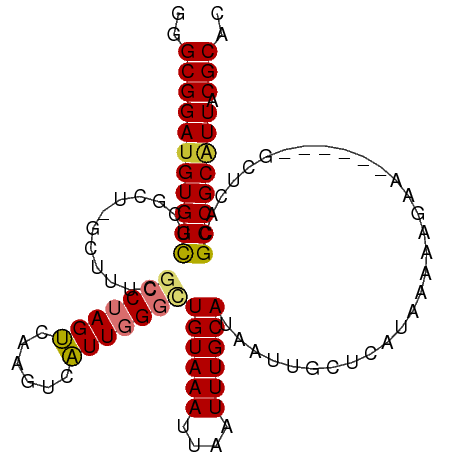
>dm3.chr3L 11173001 103 - 24543557 GGGCGGAUGUGGCUCGCUAGCUUUCGCCUAGUCAAGUCAUUGGGCUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGUAGC---UGCUCACGCCGCAUUACGCAC ..(((((((((((..((.((((..((((((((......)))))))((((((....))))))................).)))---)))....)))))))).))).. ( -32.20, z-score = -1.46, R) >droPer1.super_20 1385579 87 + 1872136 GGGCGGAUGUGGU-------------CCAAGCCGAUCCGUUCGCCUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGAA------ACUCACGCCGCGUUACGCAU (((((((((.(((-------------(......)))))))))))))............(((....)))............------.........(((...))).. ( -21.60, z-score = -0.36, R) >dp4.chrXR_group6 2781282 87 + 13314419 GGGCGGAUGUGGU-------------CCAAGCCGAUCCGUUCGCCUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGAA------ACUCACGCCGCGUUACGCAU (((((((((.(((-------------(......)))))))))))))............(((....)))............------.........(((...))).. ( -21.60, z-score = -0.36, R) >droAna3.scaffold_13337 5463485 99 - 23293914 -GGCGGAUGUGGUUUGC------UCGCCUAGUCAAGUCAUUGGGUUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGGAGCUCAUGCUCACGCCGCAUUACGCAC -.(((((((((((((((------..(((((((......))))))).))))........((((....((((........))))..))))....)))))))).))).. ( -31.20, z-score = -2.22, R) >droEre2.scaffold_4784 11182241 100 - 25762168 GGGCGGAUGUGGCUCGCUCGCUUUCGCCUAGUCAAGUCAUUGGGCUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGAA------GCUCACGCCGCAUUACGCAC ..(((((((((((......(((((.(((((((......)))))))((((((....))))))................)))------))....)))))))).))).. ( -30.10, z-score = -1.51, R) >droYak2.chr3L 11210150 100 - 24197627 GGGCGGAUGUGGCUCGCUCGCUUUCGCCUAGUCAAGUCAUUGGGCUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGAA------GCUCACGCCGCAUUACGCAC ..(((((((((((......(((((.(((((((......)))))))((((((....))))))................)))------))....)))))))).))).. ( -30.10, z-score = -1.51, R) >droSec1.super_0 3395927 103 - 21120651 GGGCGGACGUGGCUCGCUAGCUUUCGCCUAGUCAAGUCAUUGGGCUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGAAGC---UGCUCACGCCGCAUUACGCAC ..((((.(((((...((.((((((.(((((((......)))))))((((((....))))))................)))))---))))))))))))......... ( -32.00, z-score = -1.48, R) >droSim1.chr3L 10569056 103 - 22553184 GGGCGGAUGUGGCUCGCUAGCUUUCGCCUAGUCAAGUCAUUGGGCUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGAAGC---UGCUCACGCCGCAUUACGCAC ..(((((((((((..((.((((((.(((((((......)))))))((((((....))))))................)))))---)))....)))))))).))).. ( -34.40, z-score = -2.23, R) >consensus GGGCGGAUGUGGCUCGCU_GCUUUCGCCUAGUCAAGUCAUUGGGCUGUAAAUUAAUUUGCAUAAUUGCUCAUAAAAAGAA______GCUCACGCCGCAUUACGCAC ..(((((((((((............(((((((......)))))))((((((....))))))...............................)))))))).))).. (-23.23 = -23.26 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:52 2011