
| Sequence ID | dm3.chr2L |
|---|---|
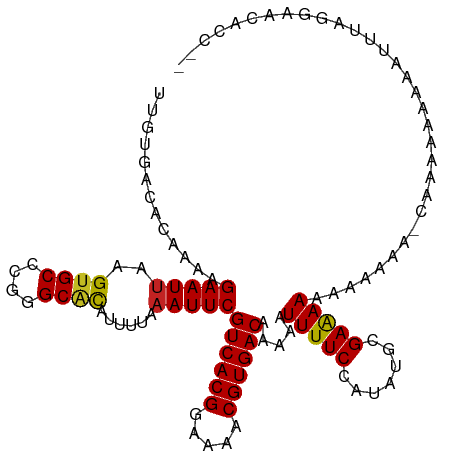
| Location | 4,805,484 – 4,805,596 |
| Length | 112 |
| Max. P | 0.995560 |

| Location | 4,805,484 – 4,805,596 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.42828 |
| G+C content | 0.35386 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -17.13 |
| Energy contribution | -16.95 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4805484 112 + 23011544 UUGUGACACAAAAGAAUGAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAAAAAAAAGAUCGGAAAAAAUUUAGGAACACC-- ..(((...............((((....)))).(((((((((.((((((.....)))))).....((((((.....................))))))))))))))).))).-- ( -25.30, z-score = -2.07, R) >droSim1.chr2L 4675212 111 + 22036055 UUGUGACACAAAAGAAUUAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAAAAAAGAU-CGGAAAAAAAUUUAGGAACACC-- ..(((...............((((....)))).(((((((((.((((((.....)))))).....((((((..................-.)))))).))))))))).))).-- ( -25.41, z-score = -2.32, R) >droSec1.super_5 2901400 111 + 5866729 UUGUGACACAAAAGAAUUAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAUAUUUUCCAUAUGCGAAAAAAAAGAU-CGGAAAAAAAUUUAGGAACACC-- ..(((...............((((....)))).(((((((((.((((((.....)))))).....((((((..................-.)))))).))))))))).))).-- ( -25.41, z-score = -2.34, R) >droYak2.chr2L 4815024 100 + 22324452 UUGUGACACAAAAGAAUUAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAAAAAA------------AAUUUAGGAACACC-- ..(((...............((((....)))).(((((((((.((((((.....)))))).....((((((....).)))))....------------))))))))).))).-- ( -23.50, z-score = -2.44, R) >droEre2.scaffold_4929 4886630 109 + 26641161 UUGUGACACAAAAGAAUUAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAAAAAAU---CCGACAAAAAUUUAGGAGCACC-- ..(((.(.............((((....)))).(((((((((.((((((.....)))))).....((((((....).))))).....---........))))))))))))).-- ( -23.40, z-score = -1.57, R) >dp4.chr4_group3 10995695 99 + 11692001 UUGUGACACAAAAGAAUUAAGUGCCCGGGCACAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAGAAGCUGAAA-CAACAUCAAC-------------- ((((................((((....))))........(((((((((.....)))))).....((((((....).)))))...))))-))).......-------------- ( -20.60, z-score = -1.48, R) >droPer1.super_8 1727739 99 - 3966273 UUGUGACACAAAAGAAUUAAGUGCCCGGGCACAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAAGCUGAAA-CAACAUCAAC-------------- ((((..((....((((((..((((....))))...........((((((.....))))))...))))))......((.....))))..)-))).......-------------- ( -20.50, z-score = -1.61, R) >droWil1.scaffold_180703 1559959 93 - 3946847 -------ACAAAAGAAUUAAGUGCCCGGGCUUAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUC-AUAUGCGAAAAAUCUGAG-AAAGAAAAACAA------------ -------......(((.(((((......))))).)))...(((((((((.....)))))).....(((((-(.((.......)).))))-)).)))......------------ ( -17.30, z-score = -1.51, R) >droVir3.scaffold_12963 10603092 112 - 20206255 GUGUGACACAAAAGAAUUAAGUGCCCAGGCACAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUC-AUAUGCGAAAAAAAAAAAACAAAACAAAAAAUGAACACAAAC- .((((...............((((....))))........(((((((((.....)))))).....(((((-......))))).....................)))))))...- ( -23.40, z-score = -3.55, R) >droMoj3.scaffold_6500 1661306 101 + 32352404 CUGUGACACAAAAGAAUUAAGUGCCCAGGCACAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUC-AUAUGCGAAAAAUAGAAA-CAGAAACGAAAAC----------- ((((................((((....))))........(((((((((.....)))))).....(((((-......)))))...))))-)))..........----------- ( -23.40, z-score = -3.81, R) >droGri2.scaffold_15252 12205530 109 + 17193109 GUGUGACACAAAAGAAUUAAGUGCC-AGGCACAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUC-AUAUGCGAAAAAGAAUA--CGAAAAAAGCAACAAACA-AAACA ((.((.(((...........))).)-).))...((((..((((((((((.....)))))).....(((((-......))))).)))).--.)))).............-..... ( -20.00, z-score = -2.06, R) >consensus UUGUGACACAAAAGAAUUAAGUGCCCGGGCACAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAAAAAAAAA_CAAAAAAAAAUUUAGGAACACC__ .............(((((..((((....))))......)))))((((((.....)))))).....(((((.......)))))................................ (-17.13 = -16.95 + -0.19)

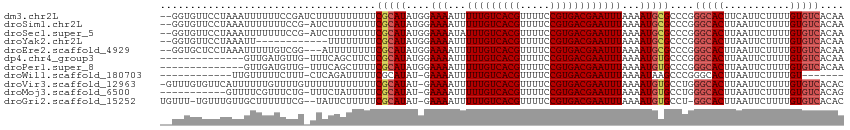
| Location | 4,805,484 – 4,805,596 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.42828 |
| G+C content | 0.35386 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -17.77 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4805484 112 - 23011544 --GGUGUUCCUAAAUUUUUUCCGAUCUUUUUUUUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUCAUUCUUUUGUGUCACAA --(((((...((((..(((((((...................)))))))..(((((((((.....)))))))))))))....)))))..(((((...........))))).... ( -24.71, z-score = -1.90, R) >droSim1.chr2L 4675212 111 - 22036055 --GGUGUUCCUAAAUUUUUUUCCG-AUCUUUUUUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAA --(((((...((((((((((((((-.................))))))).....((((((.....)))))))))))))....)))))..(((((...........))))).... ( -25.43, z-score = -2.18, R) >droSec1.super_5 2901400 111 - 5866729 --GGUGUUCCUAAAUUUUUUUCCG-AUCUUUUUUUUCGCAUAUGGAAAAUAUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAA --(((((...((((((((((((((-.................))))))).....((((((.....)))))))))))))....)))))..(((((...........))))).... ( -25.43, z-score = -2.18, R) >droYak2.chr2L 4815024 100 - 22324452 --GGUGUUCCUAAAUU------------UUUUUUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAA --(((((...((((((------------(..(((((((....))))))).....((((((.....)))))))))))))....)))))..(((((...........))))).... ( -24.30, z-score = -2.28, R) >droEre2.scaffold_4929 4886630 109 - 26641161 --GGUGCUCCUAAAUUUUUGUCGG---AUUUUUUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAA --((((((((........((.(((---(......))))))...))).....(((((((((.....)))))))))........)))))..(((((...........))))).... ( -28.30, z-score = -2.39, R) >dp4.chr4_group3 10995695 99 - 11692001 --------------GUUGAUGUUG-UUUCAGCUUCUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGUGCCCGGGCACUUAAUUCUUUUGUGUCACAA --------------(((((.....-..)))))....((((...((((....(((((((((.....)))))))))........((((....))))....))))..))))...... ( -26.60, z-score = -2.59, R) >droPer1.super_8 1727739 99 + 3966273 --------------GUUGAUGUUG-UUUCAGCUUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGUGCCCGGGCACUUAAUUCUUUUGUGUCACAA --------------(((((.....-..)))))....((((...((((....(((((((((.....)))))))))........((((....))))....))))..))))...... ( -26.60, z-score = -2.55, R) >droWil1.scaffold_180703 1559959 93 + 3946847 ------------UUGUUUUUCUUU-CUCAGAUUUUUCGCAUAU-GAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUAAGCCCGGGCACUUAAUUCUUUUGU------- ------------(((((((...((-(..(((.((((((....)-))))).))).((((((.....)))))))))...)))))))((....))...............------- ( -17.10, z-score = -1.48, R) >droVir3.scaffold_12963 10603092 112 + 20206255 -GUUUGUGUUCAUUUUUUGUUUUGUUUUUUUUUUUUCGCAUAU-GAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGUGCCUGGGCACUUAAUUCUUUUGUGUCACAC -....((((............................((((((-.......(((((((((.....)))))))))......))))))...(((((...........))))))))) ( -24.72, z-score = -2.90, R) >droMoj3.scaffold_6500 1661306 101 - 32352404 -----------GUUUUCGUUUCUG-UUUCUAUUUUUCGCAUAU-GAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGUGCCUGGGCACUUAAUUCUUUUGUGUCACAG -----------.............-............((((((-.......(((((((((.....)))))))))......))))))((((((((...........))))).))) ( -25.22, z-score = -3.86, R) >droGri2.scaffold_15252 12205530 109 - 17193109 UGUUU-UGUUUGUUGCUUUUUUCG--UAUUCUUUUUCGCAUAU-GAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGUGCCU-GGCACUUAAUUCUUUUGUGUCACAC .((((-((..........((((((--(((.(......).))))-)))))..(((((((((.....)))))))))..))))))(((..(-(((((...........))))))))) ( -24.10, z-score = -3.01, R) >consensus __GGUGUUCCUAAAUUUUUUUCUG_UUUUUUUUUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAA ....................................(((((...(((....(((((((((.....))))))))))))...)))))....(((((...........))))).... (-17.77 = -18.02 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:31 2011