
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,117,352 – 11,117,491 |
| Length | 139 |
| Max. P | 0.830075 |

| Location | 11,117,352 – 11,117,462 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 78.04 |
| Shannon entropy | 0.38346 |
| G+C content | 0.36083 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.38 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11117352 110 + 24543557 ----------UGUCAUGUAAAACGUAUUAA-AAUCGAGAGAAUAUGAAAUUCCAGUGACUCACAAGCUGGCUUCAAAUAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUGCAAAUUU- ----------.(((((......(((((...-..((....)))))))........))))).....(((((((((((.....))))((((......)))))))))))(((....)))......- ( -24.74, z-score = -1.88, R) >droEre2.scaffold_4784 11123979 99 + 25762168 ----------------------UGUUUUCCUAAUCGAGAAAAUAAAAACUUCCAGUUACUCACUAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAUUUGCAUCCUUGAAUAUUU- ----------------------...........(((((.............(((((((.....))))))).......((((((.((((......)))).)))))).....)))))......- ( -22.30, z-score = -3.01, R) >droYak2.chr3L 11145911 115 + 24197627 -----CGUUUUACCAUGUAACACGUAUUCA-AAUCUAGGGAACAGAAAUCUCCAGUUAUUCACAAGCUGGCUUUAAAAAAUGAAGCCUAACUUAAGGCGCCAUUUGCAUCCUUGAAUUUUU- -----(((.((((...)))).))).....(-(((.((((((..........((((((.......)))))).......(((((..((((......))))..)))))...)))))).))))..- ( -23.10, z-score = -1.32, R) >droSec1.super_0 3339754 120 + 21120651 UGUCUUACCUUACCAUGUAAAACGUAUUAA-AAUGGAAAGAAUAUGAAAUUCCAGUGACUCACGAGCUGGCUUCGAAAAAUGAAGCCUAACUUAAGGCGUCACUUGCAUCCUUA-AAUUUUG ..............((((((..........-..(((((...........)))))(((((.(..(((..(((((((.....)))))))...)))...).))))))))))).....-....... ( -24.20, z-score = -1.74, R) >droSim1.chr3L 10513567 115 + 22553184 -----UGUCUUACCAUGUAAAACGUAUUAA-AAUGGAGAGAAUAUGAAAUUCCAGUGACUCACAAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUG-AAUUUUG -----..((((.((((..............-.)))).))))....(((((((.((.((......(((((((((((.....))))((((......)))))))))))...)))).)-)))))). ( -27.26, z-score = -2.24, R) >consensus _________UUACCAUGUAAAACGUAUUAA_AAUCGAGAGAAUAUGAAAUUCCAGUGACUCACAAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUG_AAUUUU_ .....................................................((((((.(..(((..(((((((.....)))))))...)))...).)))))).................. (-12.86 = -13.38 + 0.52)



| Location | 11,117,352 – 11,117,462 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 78.04 |
| Shannon entropy | 0.38346 |
| G+C content | 0.36083 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -15.10 |
| Energy contribution | -16.26 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11117352 110 - 24543557 -AAAUUUGCAAGGAUGCAACUGACGCCUUAAGUUAGGCUUCAUUAUUUGAAGCCAGCUUGUGAGUCACUGGAAUUUCAUAUUCUCUCGAUU-UUAAUACGUUUUACAUGACA---------- -....(((((....))))).((((....((((((.(((((((.....)))))))))))))...))))..(((((.....))))).......-.......(((......))).---------- ( -26.10, z-score = -1.79, R) >droEre2.scaffold_4784 11123979 99 - 25762168 -AAAUAUUCAAGGAUGCAAAUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUAGUGAGUAACUGGAAGUUUUUAUUUUCUCGAUUAGGAAAACA---------------------- -...(((((((((...(....)...)))).((((.(((((((.....)))))))))))...)))))..............((((((......))))))..---------------------- ( -22.60, z-score = -1.22, R) >droYak2.chr3L 11145911 115 - 24197627 -AAAAAUUCAAGGAUGCAAAUGGCGCCUUAAGUUAGGCUUCAUUUUUUAAAGCCAGCUUGUGAAUAACUGGAGAUUUCUGUUCCCUAGAUU-UGAAUACGUGUUACAUGGUAAAACG----- -....(((((((...((((.((((((((......))))....((....)).))))..))))......((((.((.......)).)))).))-))))).(((.((((...)))).)))----- ( -23.00, z-score = -0.03, R) >droSec1.super_0 3339754 120 - 21120651 CAAAAUU-UAAGGAUGCAAGUGACGCCUUAAGUUAGGCUUCAUUUUUCGAAGCCAGCUCGUGAGUCACUGGAAUUUCAUAUUCUUUCCAUU-UUAAUACGUUUUACAUGGUAAGGUAAGACA .......-...(((....((((((..(...((((.((((((.......))))))))))...).))))))(((((.....))))).)))...-.......(((((((........))))))). ( -31.20, z-score = -2.84, R) >droSim1.chr3L 10513567 115 - 22553184 CAAAAUU-CAAGGAUGCAACUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUUGUGAGUCACUGGAAUUUCAUAUUCUCUCCAUU-UUAAUACGUUUUACAUGGUAAGACA----- .......-...(((......((((....((((((.(((((((.....)))))))))))))...))))..(((((.....))))).)))...-.......(((((((...))))))).----- ( -29.80, z-score = -2.77, R) >consensus _AAAAUU_CAAGGAUGCAAAUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUUGUGAGUCACUGGAAUUUCAUAUUCUCUCGAUU_UUAAUACGUUUUACAUGGUAA_________ ....................((((....((((((.(((((((.....)))))))))))))...))))..(((((.....)))))...................................... (-15.10 = -16.26 + 1.16)

| Location | 11,117,371 – 11,117,491 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Shannon entropy | 0.34686 |
| G+C content | 0.34868 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -16.34 |
| Energy contribution | -17.50 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
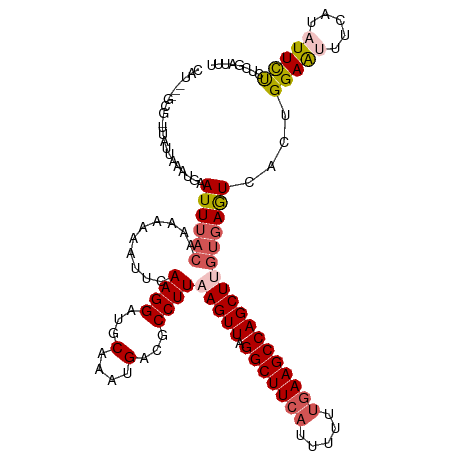
>dm3.chr3L 11117371 120 - 24543557 CAUAGAGCGCUUAUUAAAUGAAUUUACAAAAAUUUGCAAGGAUGCAACUGACGCCUUAAGUUAGGCUUCAUUAUUUGAAGCCAGCUUGUGAGUCACUGGAAUUUCAUAUUCUCUCGAUUU ....(((.(..(((.((((..............(((((....))))).((((....((((((.(((((((.....)))))))))))))...)))).....)))).)))..).)))..... ( -28.80, z-score = -1.53, R) >droEre2.scaffold_4784 11123988 119 - 25762168 CAUGAGGCGUUUACUAAAAGUAUUUACUAAAAUAUUCAAGGAUGCAAAUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUAGUGAGUAACUGGAAGUUUUUAUUUUCUCGAUU- ..(((((((((........((((((..............))))))....)))))))))((((.(((((((.....)))))))))))(((.....)))((((((....))))))......- ( -29.24, z-score = -1.96, R) >droYak2.chr3L 11145935 120 - 24197627 CAUAAAGCGGUUAUUAAAAGUAUUAAGUAAAAAAUUCAAGGAUGCAAAUGGCGCCUUAAGUUAGGCUUCAUUUUUUAAAGCCAGCUUGUGAAUAACUGGAGAUUUCUGUUCCCUAGAUUU (((((.((((((.(((((((.((.((((....((((.((((.(((.....))))))).))))..)))).))))))))))))).))))))).....((((.((.......)).)))).... ( -25.80, z-score = -0.75, R) >droSec1.super_0 3339783 109 - 21120651 -----------CACUAAAUGAAUUUCCAACAAAAUUUAAGGAUGCAAGUGACGCCUUAAGUUAGGCUUCAUUUUUCGAAGCCAGCUCGUGAGUCACUGGAAUUUCAUAUUCUUUCCAUUU -----------......(((((.(((((....(((((((((...(....)...))))))))).((((((.......))))))..............))))).)))))............. ( -28.20, z-score = -3.10, R) >droSim1.chr3L 10513591 109 - 22553184 -----------CAUUAAAUGAAUUUACAACAAAAUUCAAGGAUGCAACUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUUGUGAGUCACUGGAAUUUCAUAUUCUCUCCAUUU -----------.......(((((((......))))))).(((......((((....((((((.(((((((.....)))))))))))))...))))..(((((.....))))).))).... ( -29.00, z-score = -3.32, R) >consensus CAU___GCG_UUAUUAAAUGAAUUUACAAAAAAAUUCAAGGAUGCAAAUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUUGUGAGUCACUGGAAUUUCAUAUUCUCUCGAUUU .....................((((((..........((((...(....)...))))(((((.(((((((.....))))))))))))))))))....(((((.....)))))........ (-16.34 = -17.50 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:48 2011