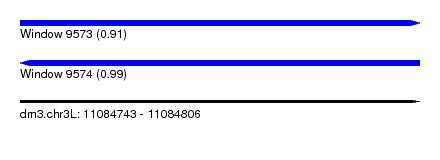
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,084,743 – 11,084,806 |
| Length | 63 |
| Max. P | 0.993104 |

| Location | 11,084,743 – 11,084,806 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 64.23 |
| Shannon entropy | 0.69323 |
| G+C content | 0.41575 |
| Mean single sequence MFE | -9.67 |
| Consensus MFE | -4.50 |
| Energy contribution | -4.44 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
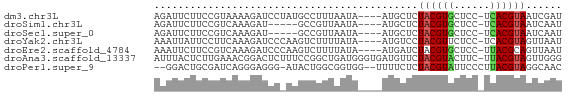
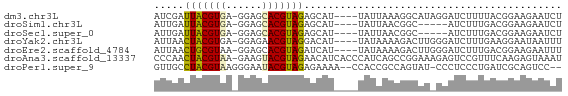
>dm3.chr3L 11084743 63 + 24543557 AGAUUCUUCCGUAAAAGAUCCUAUGCCUUUAAUA----AUGCUCUACGUGCUCC-UCACGUAAUCGAU ((..((((......))))..))............----......((((((....-.))))))...... ( -7.30, z-score = -0.79, R) >droSim1.chr3L 10480335 58 + 22553184 AGAUUCUUCCGUCAAAGAU-----GCCGUUAAUA----AUGCUCUACGUGCUCC-UCACGUAAUCAAU .((((..........(((.-----((........----..)))))(((((....-.)))))))))... ( -6.70, z-score = -0.50, R) >droSec1.super_0 3310384 58 + 21120651 AGAUUCUUCCGUCAAAGAU-----GCCGUUAAUA----AUGCUCUACGUGCUCC-UCACGUAAUCAAU .((((..........(((.-----((........----..)))))(((((....-.)))))))))... ( -6.70, z-score = -0.50, R) >droYak2.chr3L 11115395 63 + 24197627 AAAUUAUUCCUUCAAAGAUCCCAAGUCUUUUAUA----AUGUCCUACGUUCUCC-UCACGUAGUUAAU .............((((((.....))))))....----.....((((((.....-..))))))..... ( -7.50, z-score = -3.56, R) >droEre2.scaffold_4784 11093511 63 + 25762168 AAAUUCUUCCGUCAAAGAUCCCAAGUCUUUUAUA----AUGAUCUACGUGCUCC-UUACGCAGUUAAU ..........((.((((((.....)))))).)).----.(((.((.((((....-.)))).))))).. ( -5.00, z-score = -0.30, R) >droAna3.scaffold_13337 8803695 67 - 23293914 AUUUACUCUUGAAACGGACUCUUUCCGGCUGAUGGGUGAUGUUCUACGUACUUC-UUACGUAGUUGGG ..((((((.(.(..((((.....))))..).).))))))....(((((((....-.)))))))..... ( -17.20, z-score = -1.82, R) >droPer1.super_9 2021025 63 + 3637205 --GGACUGCGAUCAGGGAGGG-AUACUGGCGGUGG--UUUUCUCUACGUAUUCCCUUACGUAGGCAAC --...(((((......(((((-(((((((.((...--...)).))).))).)))))).)))))..... ( -17.30, z-score = -0.37, R) >consensus AGAUUCUUCCGUCAAAGAUCC_AUGCCGUUAAUA____AUGCUCUACGUGCUCC_UCACGUAGUCAAU ............................................((((((......))))))...... ( -4.50 = -4.44 + -0.06)
| Location | 11,084,743 – 11,084,806 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 64.23 |
| Shannon entropy | 0.69323 |
| G+C content | 0.41575 |
| Mean single sequence MFE | -10.99 |
| Consensus MFE | -7.57 |
| Energy contribution | -7.24 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
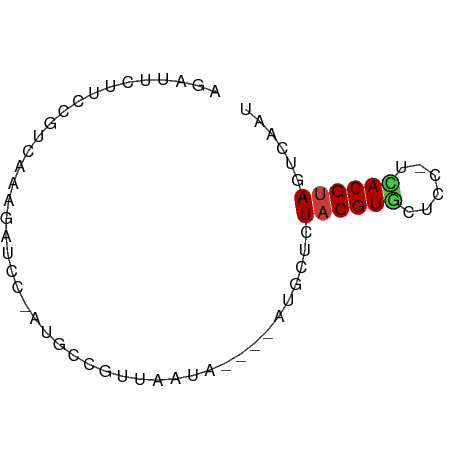
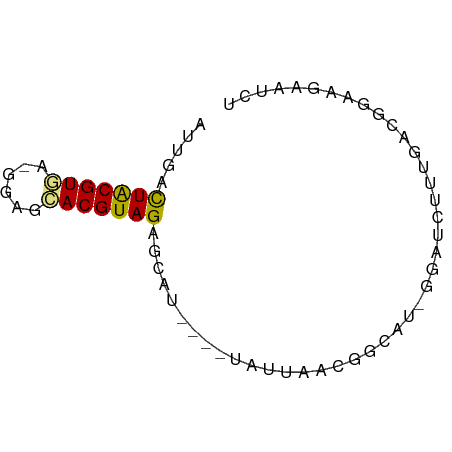
>dm3.chr3L 11084743 63 - 24543557 AUCGAUUACGUGA-GGAGCACGUAGAGCAU----UAUUAAAGGCAUAGGAUCUUUUACGGAAGAAUCU .....(((((((.-....))))))).((..----........))...((((((((....)))).)))) ( -10.60, z-score = -0.61, R) >droSim1.chr3L 10480335 58 - 22553184 AUUGAUUACGUGA-GGAGCACGUAGAGCAU----UAUUAACGGC-----AUCUUUGACGGAAGAAUCU ...((((.(((((-(((((.(((.((....----..)).)))))-----.))))).)))....)))). ( -13.10, z-score = -1.83, R) >droSec1.super_0 3310384 58 - 21120651 AUUGAUUACGUGA-GGAGCACGUAGAGCAU----UAUUAACGGC-----AUCUUUGACGGAAGAAUCU ...((((.(((((-(((((.(((.((....----..)).)))))-----.))))).)))....)))). ( -13.10, z-score = -1.83, R) >droYak2.chr3L 11115395 63 - 24197627 AUUAACUACGUGA-GGAGAACGUAGGACAU----UAUAAAAGACUUGGGAUCUUUGAAGGAAUAAUUU .....((((((..-.....)))))).....----....((((((....).)))))............. ( -8.90, z-score = -0.95, R) >droEre2.scaffold_4784 11093511 63 - 25762168 AUUAACUGCGUAA-GGAGCACGUAGAUCAU----UAUAAAAGACUUGGGAUCUUUGACGGAAGAAUUU .....((((((..-.....)))))).....----....((((((....).)))))..(....)..... ( -8.80, z-score = -0.17, R) >droAna3.scaffold_13337 8803695 67 + 23293914 CCCAACUACGUAA-GAAGUACGUAGAACAUCACCCAUCAGCCGGAAAGAGUCCGUUUCAAGAGUAAAU .....(((((((.-....)))))))................((((.....)))).............. ( -11.50, z-score = -2.35, R) >droPer1.super_9 2021025 63 - 3637205 GUUGCCUACGUAAGGGAAUACGUAGAGAAAA--CCACCGCCAGUAU-CCCUCCCUGAUCGCAGUCC-- .(((((((((((......)))))))......--.......(((...-......)))...))))...-- ( -10.90, z-score = -0.17, R) >consensus AUUGACUACGUGA_GGAGCACGUAGAGCAU____UAUUAACGGCAU_GGAUCUUUGACGGAAGAAUCU .....(((((((......)))))))........................................... ( -7.57 = -7.24 + -0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:41 2011