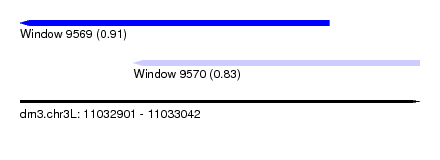
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,032,901 – 11,033,042 |
| Length | 141 |
| Max. P | 0.907321 |

| Location | 11,032,901 – 11,033,010 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.51 |
| Shannon entropy | 0.53407 |
| G+C content | 0.50686 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -18.20 |
| Energy contribution | -17.83 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
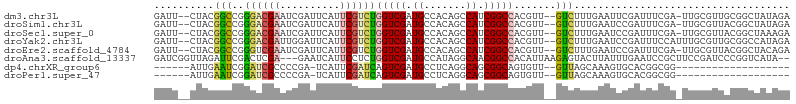
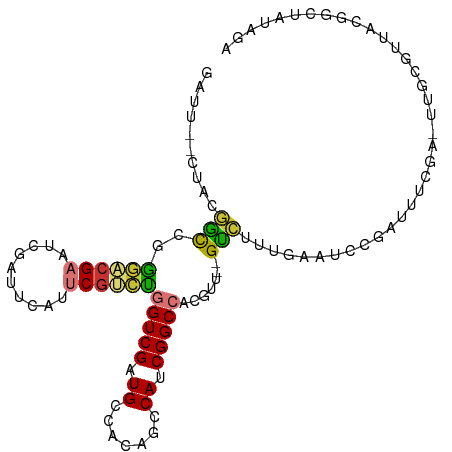
>dm3.chr3L 11032901 109 - 24543557 UCUGGUCGAUGCCACAGCCAUCGGCCACGUUGUCUUUGAAUUCGAUUUCGA-UUGCGUUGCGGCUAUAGAAACUCGAUCGCGAUGUACAAGAACAUCGACAAUGAGUCCU ..((((((((((....).)))))))))(((((((...(...((((((((..-..((......))....)))).))))...)(((((......))))))))))))...... ( -34.90, z-score = -1.75, R) >droSim1.chr3L 10430579 109 - 22553184 UCUGGUCGAUGCCACAGCCAUCGGCCACGUUGUCUUUGAAUCCGAUUUCGA-UUGCGUUACGGCUAUAGAAACUCGAUCGCGAUGUACAAGAACAUCGACAACGAGUCCU ..((((((((((....).)))))))))(((((((..............(((-(((.(((.(.......).))).)))))).(((((......))))))))))))...... ( -35.50, z-score = -2.28, R) >droSec1.super_0 3260634 109 - 21120651 UCUGGUCGAUGCCACAGCCAUCGGCCACGUUGUCUUUGAAUCCGAUUUCGA-UUGCGUUACGGCUAAAGAAACCCGAUCGCGAUGUACAAGAACAUCGACAACGAGUCCU ..((((((((((....).)))))))))(((((((.(((((......)))))-..(((...(((..........)))..)))(((((......))))))))))))...... ( -35.50, z-score = -2.49, R) >droYak2.chr3L 11061155 110 - 24197627 UCUGGUCGAUGCCACAGCCAUCGGCCACGUUGUCUUUGAAUCCGAUUUCCAUUUGCGUUGCGGCCAUAGAAACUCGAUCGCGAUGUACAAGAACAUCGACAACGAGUCCU ..((((((((((....).)))))))))(((((((...(....)((((((....(((((((((..(..........)..)))))))))...))).))))))))))...... ( -34.70, z-score = -1.94, R) >droEre2.scaffold_4784 11041748 109 - 25762168 UCUGGUCGAUGCCACAGCCAUCGGCCACGUUGUCUUUGAAUCCGAUUUCGA-UUGCGUUACGGCUACAGAAUCACGAUCGCGAUGUACAAGAACAUCGACAACGAGUCCU ..((((((((((....).)))))))))(((((((.(((((......)))))-..(((...((............))..)))(((((......))))))))))))...... ( -33.70, z-score = -1.53, R) >droAna3.scaffold_13337 15946212 102 - 23293914 UCUGGUCGAUGCCAUAGGCAACGGCCACAUUA----AGAGUACUUAUUUGAAUCCGCUUCCGAUCCCGG----UCAUACGCGAUGUACAAGAACAUUGACAAUGAGUCCU ..((((((.((((...)))).)))))).....----..((.(((((((...........(((....)))----......(((((((......)))))).)))))))).)) ( -25.80, z-score = -1.03, R) >dp4.chrXR_group6 7323006 83 + 13314419 AUCAGUCGAUGCCUCAGGCAGCGGCAGUGUUGUUAGCAAA------------GUGCACGGCGG---------------CGCGAUGUACAAGAACAUUGACAACGAGUCCU .((.((((.((((...)))).))))...((((((......------------((((......)---------------)))(((((......)))))))))))))..... ( -24.40, z-score = 0.07, R) >droPer1.super_47 100919 83 + 592741 AUCAGUCGAUGCCUCAGGCAGCGGCAGUGUUGUUAGCAAA------------GUGCACGGCGG---------------CGCGAUGUACAAGAACAUUGACAACGAGUCCU .((.((((.((((...)))).))))...((((((......------------((((......)---------------)))(((((......)))))))))))))..... ( -24.40, z-score = 0.07, R) >consensus UCUGGUCGAUGCCACAGCCAUCGGCCACGUUGUCUUUGAAUCCGAUUUCGA_UUGCGUUGCGGCUAUAGAAACUCGAUCGCGAUGUACAAGAACAUCGACAACGAGUCCU ..((((((.((.......)).))))))(((((((....................((.......................))(((((......))))))))))))...... (-18.20 = -17.83 + -0.37)

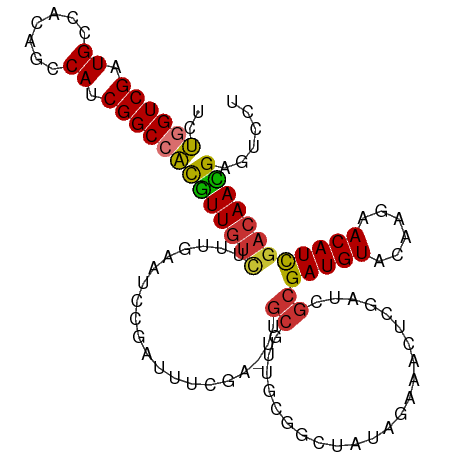
| Location | 11,032,941 – 11,033,042 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 66.50 |
| Shannon entropy | 0.64986 |
| G+C content | 0.52527 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -10.82 |
| Energy contribution | -10.16 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.833888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11032941 101 - 24543557 GAUU--CUACGGCCGGGACGAAUCGAUUCAUUCGUCUGGUCGAUGCCACAGCCAUCGGCCACGUU--GUCUUUGAAUUCGAUUUCGA-UUGCGUUGCGGCUAUAGA ...(--(((..((((.((((((((((((((..((.(((((((((((....).))))))))).).)--)....)))).......))))-)).)))).))))..)))) ( -38.71, z-score = -2.68, R) >droSim1.chr3L 10430619 101 - 22553184 GAUU--CUACGGCCGGGACGAAUCGAUUCAUUCGUCUGGUCGAUGCCACAGCCAUCGGCCACGUU--GUCUUUGAAUCCGAUUUCGA-UUGCGUUACGGCUAUAGA ...(--(((..((((..(((((((((((((..((.(((((((((((....).))))))))).).)--)....)))))).(....)))-)).)))..))))..)))) ( -38.60, z-score = -2.93, R) >droSec1.super_0 3260674 101 - 21120651 GAUU--CUACGGCCGGGACGAAUCGAUUCAUUCGUCUGGUCGAUGCCACAGCCAUCGGCCACGUU--GUCUUUGAAUCCGAUUUCGA-UUGCGUUACGGCUAAAGA ...(--((..(((((..(((((((((((((..((.(((((((((((....).))))))))).).)--)....)))))).(....)))-)).)))..)))))..))) ( -35.70, z-score = -2.05, R) >droYak2.chr3L 11061195 102 - 24197627 GAUU--CUACGGCCGGGACGAUUGGAUUCAUUCGUCUGGUCGAUGCCACAGCCAUCGGCCACGUU--GUCUUUGAAUCCGAUUUCCAUUUGCGUUGCGGCCAUAGA ...(--(((.(((((.((((((((((((((..((.(((((((((((....).))))))))).).)--)....)))))))))).........)))).))))).)))) ( -43.10, z-score = -3.90, R) >droEre2.scaffold_4784 11041788 101 - 25762168 GAUU--CUACGGCCGGGUCGAAUCGAUUCAUUCGUCUGGUCGAUGCCACAGCCAUCGGCCACGUU--GUCUUUGAAUCCGAUUUCGA-UUGCGUUACGGCUACAGA ...(--((..((((((((((((((((((((..((.(((((((((((....).))))))))).).)--)....))))).))).)))))-))......)))))..))) ( -33.60, z-score = -1.32, R) >droAna3.scaffold_13337 15946244 101 - 23293914 GAUCGGUUAGAUUCGACUCGA---GAAUCAUUCCUCUGGUCGAUGCCAUAGGCAACGGCCACAUUAAGAGUACUUAUUUGAAUCCGCUUCCGAUCCCGGUCAUA-- ((((((...((((((((((((---(........)))((((((.((((...)))).))))))......)))).......)))))).....)))))).........-- ( -32.21, z-score = -2.56, R) >dp4.chrXR_group6 7323038 78 + 13314419 ------AUUGAAUCGGAUCGCCCCGA-UCAUUCGAUCAGUCGAUGCCUCAGGCAGCGGCAGUGUU--GUUAGCAAAGUGCACGGCGG------------------- ------............((((..((-((....)))).((((.((((...)))).)))).((((.--...........)))))))).------------------- ( -23.50, z-score = -0.04, R) >droPer1.super_47 100951 78 + 592741 ------AUUGAAUCGGAUCGCCCCGA-UCAUUCGAUCAGUCGAUGCCUCAGGCAGCGGCAGUGUU--GUUAGCAAAGUGCACGGCGG------------------- ------............((((..((-((....)))).((((.((((...)))).)))).((((.--...........)))))))).------------------- ( -23.50, z-score = -0.04, R) >consensus GAUU__CUACGGCCGGGACGAAUCGAUUCAUUCGUCUGGUCGAUGCCACAGCCAUCGGCCACGUU__GUCUUUGAAUCCGAUUUCGA_UUGCGUUACGGCUAUAGA ..........(((..((((((..........))))))(((((.((.......)).))))).......))).................................... (-10.82 = -10.16 + -0.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:38 2011