| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,801,986 – 4,802,079 |
| Length | 93 |
| Max. P | 0.717979 |

| Location | 4,801,986 – 4,802,079 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 62.41 |
| Shannon entropy | 0.79692 |
| G+C content | 0.60131 |
| Mean single sequence MFE | -39.54 |
| Consensus MFE | -10.61 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.717979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4801986 93 - 23011544 UUGCUGCUGCCAAAGCUUC-GCUGCAGCAGCAGCUGCGGUCAGCGU-----CGCCGUCGCCGCUGGC-GUCGCAGUCGCAGCUUUUUCAAAGCUGCACGA .(((((((((...(((...-))))))))))))(((((((((((((.-----((....)).)))))))-..)))))).(((((((.....))))))).... ( -48.50, z-score = -2.59, R) >droSim1.chr2L 4671766 93 - 22036055 UUGCUGCUGCCAAAGCUUC-GCUGCAGCAGCAGCUGCGGUCAGCGU-----CGCCGUCGCCGCUGGC-GUCGCAGUCGCAGCUUUUUCAAAGCUGCACGA .(((((((((...(((...-))))))))))))(((((((((((((.-----((....)).)))))))-..)))))).(((((((.....))))))).... ( -48.50, z-score = -2.59, R) >droSec1.super_5 2898007 93 - 5866729 UCGCUGCUGCCAAAGCUUC-GCUGCAGCAGCAGCUGCGGUCAGCGU-----CGCCGUCGCCGCUGGC-GUCGCAGUCGCAGCUUUUUCAAAGCUGCACGA ..((((((((...(((...-))))))))))).(((((((((((((.-----((....)).)))))))-..)))))).(((((((.....))))))).... ( -47.90, z-score = -2.55, R) >droYak2.chr2L 4811397 92 - 22324452 UUGCUGCUGCCAAAGCUUC-GCUGCAGCAGAAGUUGCGGUCAGCGU-----CGCCGUUGGCGUUGGC-GUCGCAGUCGCAGC-UUUUCAAAGCUGCACGA ..(((((((((((.(((..-((((((((....)))))))).)))..-----((((...)))))))))-)..))))).(((((-((....))))))).... ( -42.90, z-score = -1.65, R) >droEre2.scaffold_4929 4883132 92 - 26641161 UUGCUGCUGCCAAAGCUUC-GCUGCAGCAGAAGCUGCGGUCAGCUU-----CACCGCCGCCGUUGGC-GUCGCCGUCGCAGC-UUUUCAAAGCUGCACGA .((((((.((.........-)).))))))(((((((....))))))-----)..((.((((...)))-).)).(((.(((((-((....)))))))))). ( -40.00, z-score = -1.21, R) >droAna3.scaffold_12916 9369927 94 - 16180835 CUUUUGCUGGUAAAGCUUCGGCUGCAGCCUCUGUUGCUGCCUCAGU-----CGACGUUGGCAGUGGCAGUCGACGUCGCAGC-UUUUCAAAGCUGCACGA .....((((....(((....))).))))..((((..(((((...(.-----...)...)))))..))))....(((.(((((-((....)))))))))). ( -41.70, z-score = -2.70, R) >droWil1.scaffold_180703 1555292 71 + 3946847 ---------------UUGC-ACUGCCUUAAC--UUGGCUUUAGCUUUUCGAAGCCGACUUCGUUGC--CGAGCCACUUCAGAGUUGGCCAA--------- ---------------....-...(((..(((--(((((((..((....(((((....)))))..))--.))))))......)))))))...--------- ( -18.00, z-score = -0.37, R) >droMoj3.scaffold_6500 12218331 82 + 32352404 -UGAUCUUGACGAAGCU---GCCACGUGAGCAGCUGCAGCCAGCGU-----UGCGAGAGAUUGUGGG-AUUCCAGUUGCAGC---CGCUAGUCCA----- -.((((((.....((((---(((....).))))))(((((....))-----)))..))))))...((-(((..(((......---.)))))))).----- ( -23.80, z-score = 0.91, R) >anoGam1.chr2R 48649092 99 + 62725911 UUGCUGCUGAUCACGCUGC-GGUGCAGCAGCAGCAGCUGUUUGCGCCCCGGCACCACUGUUACCGGACGGUGCAACCGCAGCAGUGACCGGACUGGAAGA ((.(..(((.(((((((((-(((..((((((....))))))((((((((((...........))))..)))))))))))))).)))).)))...).)).. ( -44.60, z-score = -1.46, R) >consensus UUGCUGCUGCCAAAGCUUC_GCUGCAGCAGCAGCUGCGGUCAGCGU_____CGCCGUCGCCGCUGGC_GUCGCAGUCGCAGC_UUUUCAAAGCUGCACGA ....(((((...............)))))...(((((((.(.(((......((.((....)).)).....))).)))))))).................. (-10.61 = -10.75 + 0.14)

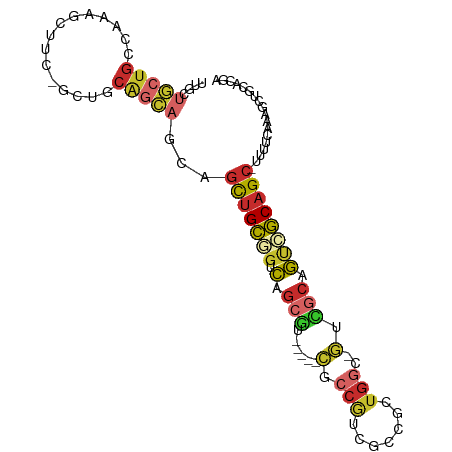

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:29 2011