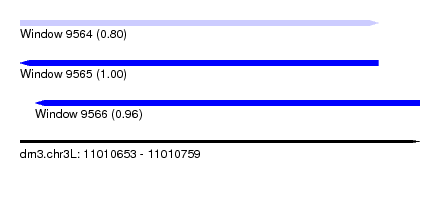
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,010,653 – 11,010,759 |
| Length | 106 |
| Max. P | 0.999277 |

| Location | 11,010,653 – 11,010,748 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.05 |
| Shannon entropy | 0.37008 |
| G+C content | 0.38409 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -10.76 |
| Energy contribution | -11.64 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
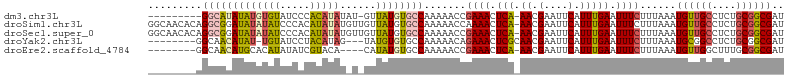
>dm3.chr3L 11010653 95 + 24543557 ---------GGCAUAUAUGUGUAUCCCACAUAUAU-GUUAUGUGCCAAAAACCGAAACUCA-AACGAAUUCAUUUGAAUUUCUUUAAAUGUUGCCUCUGCGGCGAU ---------(((((((((((((((.....))))))-).)))))))).......((((.(((-((.(....).))))).)))).......((((((.....)))))) ( -25.30, z-score = -3.61, R) >droSim1.chr3L 10407416 105 + 22553184 GGCAACACAGGCGGAUAUAUAUCCCACAUAUAUGUUGUUAUGUGCCAAAAACCAAAACUCA-AACGAAUUCAUUUGAAUUUCUUUAAAUGUUGCCUCUGCGGCGAU (((((((..(((((((....)))).(((((........))))))))..............(-((.((((((....)))))).)))...)))))))........... ( -25.50, z-score = -2.26, R) >droSec1.super_0 3233954 105 + 21120651 GGCAACACAGGCGGAUAUAUAUCCCACAUAUAUGUUGUUAUGUGCCAAAAACCGAAACUCA-AACGAAUUCAUUUGAAUUUCUUUAAAUGUUGCCUCUGCGGCGAU (((((((..(((((((....)))).(((((........)))))))).......((((.(((-((.(....).))))).))))......)))))))........... ( -27.80, z-score = -2.77, R) >droYak2.chr3L 11038294 94 + 24197627 --------GGCAACAUAU-UGUAUCCUACAUAG---UAUGUGUGCCAAAAACAGAAACUCGCAACGAAUUCAUUUGAAUUUCUUUAAAUGCGGCCUCUGCGGCGAU --------((((((((((-(((.......))))---))))).))))............((((...((((((....)))))).......(((((...))))))))). ( -24.50, z-score = -2.37, R) >droEre2.scaffold_4784 11018642 93 + 25762168 --------GGCAACAUGCACAUAUAUCGUACA----CAUAUGUGCCAAAAACCGAAACUCA-AACGAAUUCAUUUGAAUUUCUUUAAAUGUUGGCUUUGCGGCGAU --------(.((((((((((((((........----.))))))))........((((.(((-((.(....).))))).)))).....)))))).)........... ( -21.30, z-score = -1.90, R) >consensus ________AGGCACAUAUAUAUAUCACAUAUAU_U_GUUAUGUGCCAAAAACCGAAACUCA_AACGAAUUCAUUUGAAUUUCUUUAAAUGUUGCCUCUGCGGCGAU .........(((((((((((((.....)))))......))))))))...................((((((....)))))).......((((((....)))))).. (-10.76 = -11.64 + 0.88)
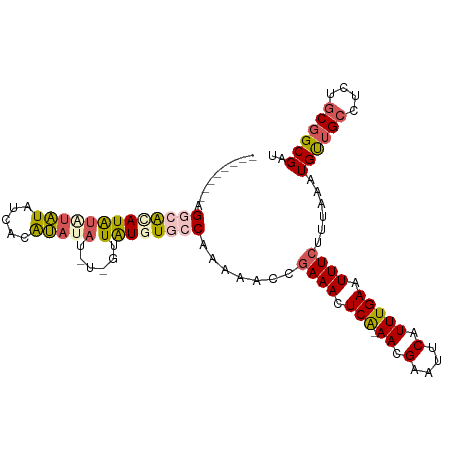

| Location | 11,010,653 – 11,010,748 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.05 |
| Shannon entropy | 0.37008 |
| G+C content | 0.38409 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -17.16 |
| Energy contribution | -16.28 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.27 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11010653 95 - 24543557 AUCGCCGCAGAGGCAACAUUUAAAGAAAUUCAAAUGAAUUCGUU-UGAGUUUCGGUUUUUGGCACAUAAC-AUAUAUGUGGGAUACACAUAUAUGCC--------- ...((((.((((....).......((((((((((((....))))-))))))))..))).))))......(-(((((((((.....))))))))))..--------- ( -33.20, z-score = -5.75, R) >droSim1.chr3L 10407416 105 - 22553184 AUCGCCGCAGAGGCAACAUUUAAAGAAAUUCAAAUGAAUUCGUU-UGAGUUUUGGUUUUUGGCACAUAACAACAUAUAUGUGGGAUAUAUAUCCGCCUGUGUUGCC ...((.(((.((((..((......((((((((((((....))))-))))))))......)).((((((........))))))((((....)))))))).))).)). ( -34.90, z-score = -4.13, R) >droSec1.super_0 3233954 105 - 21120651 AUCGCCGCAGAGGCAACAUUUAAAGAAAUUCAAAUGAAUUCGUU-UGAGUUUCGGUUUUUGGCACAUAACAACAUAUAUGUGGGAUAUAUAUCCGCCUGUGUUGCC ...((.(((.((((..((......((((((((((((....))))-))))))))......)).((((((........))))))((((....)))))))).))).)). ( -36.70, z-score = -4.72, R) >droYak2.chr3L 11038294 94 - 24197627 AUCGCCGCAGAGGCCGCAUUUAAAGAAAUUCAAAUGAAUUCGUUGCGAGUUUCUGUUUUUGGCACACAUA---CUAUGUAGGAUACA-AUAUGUUGCC-------- ......((((((((((((......(((.(((....))))))..)))).))))))))....((((.(((((---...((((...))))-.)))))))))-------- ( -24.80, z-score = -1.82, R) >droEre2.scaffold_4784 11018642 93 - 25762168 AUCGCCGCAAAGCCAACAUUUAAAGAAAUUCAAAUGAAUUCGUU-UGAGUUUCGGUUUUUGGCACAUAUG----UGUACGAUAUAUGUGCAUGUUGCC-------- ...((......))((((((.....((((((((((((....))))-))))))))........(((((((((----(.....))))))))))))))))..-------- ( -30.70, z-score = -4.01, R) >consensus AUCGCCGCAGAGGCAACAUUUAAAGAAAUUCAAAUGAAUUCGUU_UGAGUUUCGGUUUUUGGCACAUAAC_A_AUAUAUGGGAUAUAUAUAUCUGCCC________ ...((((.((((....).......((((((((((((....)))).))))))))..))).))))...........((((((.....))))))............... (-17.16 = -16.28 + -0.88)
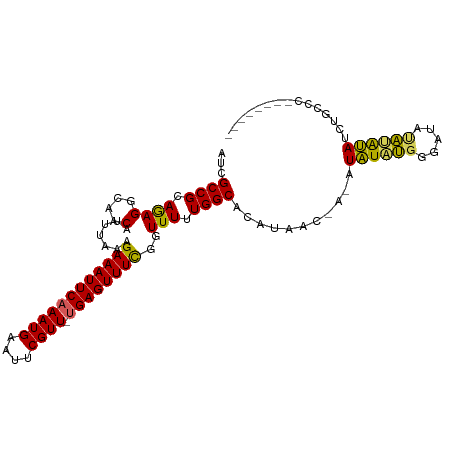
| Location | 11,010,657 – 11,010,759 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 61.48 |
| Shannon entropy | 0.73761 |
| G+C content | 0.35111 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -2.25 |
| Energy contribution | -2.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.09 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
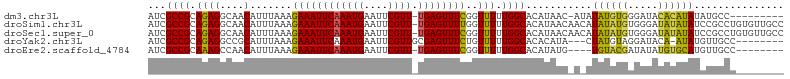
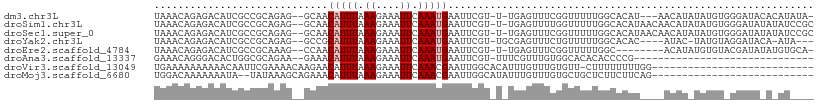
>dm3.chr3L 11010657 102 - 24543557 UAAACAGAGACAUCGCCGCAGAG--GCAACAUUUAAAGAAAUUCAAAUGAAUUCGU-U-UGAGUUUCGGUUUUUGGCACAU---AACAUAUAUGUGGGAUACACAUAUA- ....(((((((...(((.....)--))..........((((((((((((....)))-)-)))))))).)))))))......---....((((((((.....))))))))- ( -33.80, z-score = -6.72, R) >droSim1.chr3L 10407426 106 - 22553184 UAAACAGAGACAUCGCCGCAGAG--GCAACAUUUAAAGAAAUUCAAAUGAAUUCGU-U-UGAGUUUUGGUUUUUGGCACAUAACAACAUAUAUGUGGGAUAUAUAUCCGC ....(((((((...(((.....)--))..........((((((((((((....)))-)-)))))))).))))))).((((((........))))))((((....)))).. ( -31.90, z-score = -4.97, R) >droSec1.super_0 3233964 106 - 21120651 UAAACAGAGACAUCGCCGCAGAG--GCAACAUUUAAAGAAAUUCAAAUGAAUUCGU-U-UGAGUUUCGGUUUUUGGCACAUAACAACAUAUAUGUGGGAUAUAUAUCCGC ....(((((((...(((.....)--))..........((((((((((((....)))-)-)))))))).))))))).((((((........))))))((((....)))).. ( -34.40, z-score = -5.87, R) >droYak2.chr3L 11038301 98 - 24197627 UAAACAGAGACAUCGCCGCAGAG--GCCGCAUUUAAAGAAAUUCAAAUGAAUUCGU-UGCGAGUUUCUGUUUUUGGCACAC----AUAC-UAUGUAGGAUACA-AUA--- .((((((((((.((((.((.(((--....(((((.((....)).)))))..)))))-.))))))))))))))(((.(..((----((..-.))))..)...))-)..--- ( -25.20, z-score = -2.57, R) >droEre2.scaffold_4784 11018649 97 - 25762168 UAAACAGAGACAUCGCCGCAAAG--CCAACAUUUAAAGAAAUUCAAAUGAAUUCGU-U-UGAGUUUCGGUUUUUGGC--------ACAUAUGUGUACGAUAUAUGUGCA- ....(((((((...((......)--)...........((((((((((((....)))-)-)))))))).)))))))((--------((((((((.....)))))))))).- ( -33.30, z-score = -5.37, R) >droAna3.scaffold_13337 15923143 77 - 23293914 GAAACAGGGACACUGGCGCAGAA--GAAACAUUUAAAGAAAUUCAAAUGAAUUCGU-UUUCGUUUGUGGCACACACCCCG------------------------------ ......(((....((.((((((.--(((((((((.((....)).))))).......-)))).)))))).)).....))).------------------------------ ( -16.71, z-score = -1.17, R) >droVir3.scaffold_13049 15683310 82 - 25233164 UGAAAAAAAAAACAAUUCGAAAACAAGAACAUUUAAAGAAAUUCAAACGAAUUGGCACAUUUGUUUGUGUU-CUUUUUUUUGG--------------------------- .................((((((.(((((((....((....))(((((((((......)))))))))))))-))).)))))).--------------------------- ( -15.90, z-score = -1.45, R) >droMoj3.scaffold_6680 3612638 81 + 24764193 UGGACAAAAAAAUA--UAUAAAGCAGAAACAUUUAAAGAAAUUCAAACGAAUUGGCAUAUUUGUUUGUGCUGCUCUUCUUCAG--------------------------- ..............--.....((((.(((((..((....(((((....)))))....))..))))).))))............--------------------------- ( -13.90, z-score = -1.09, R) >consensus UAAACAGAGACAUCGCCGCAGAG__GCAACAUUUAAAGAAAUUCAAAUGAAUUCGU_U_UGAGUUUCGGUUUUUGGCACAU____ACAU_UAUGU_GGAUA_A_AU____ .............................(((((.((....)).)))))((((((....))))))............................................. ( -2.25 = -2.50 + 0.25)
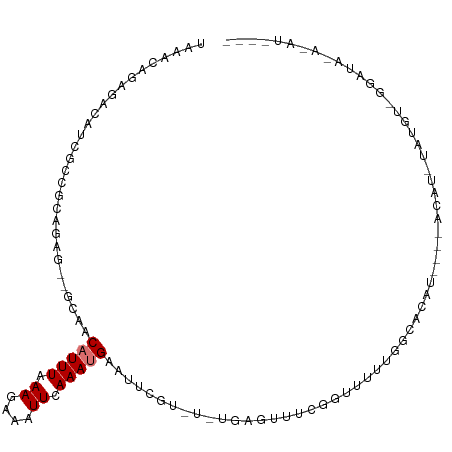

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:34 2011