
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,966,179 – 10,966,271 |
| Length | 92 |
| Max. P | 0.639439 |

| Location | 10,966,179 – 10,966,271 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 67.34 |
| Shannon entropy | 0.64590 |
| G+C content | 0.63111 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -11.85 |
| Energy contribution | -12.23 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10966179 92 + 24543557 ACCACUACGCUCGCCUGCACUCCGCUCCGGUGGUGCACCAUCAUCACCAUGACACCCACGCCGCCGUGGUG-CAUCACAGUACCCCGGCCCAC------ ........((......))..........(((((((......)))))))..............((((.((((-(......))))).))))....------ ( -26.50, z-score = -0.73, R) >droSim1.chr3L 10361199 92 + 22553184 ACCACUACGCUCGCCUGCACUCCGCUCCGGUGGUGCACCACCAUCACCAUGACACCCAUGCCGCCGUGGUG-CAUCACAGUAUCCCGGCUCAC------ ............(((.((.....))....((((((((((((......((((.....)))).....))))))-))))))........)))....------ ( -30.00, z-score = -1.89, R) >droSec1.super_0 3189821 92 + 21120651 ACCACUACGCUCGCCUGCACUCCGCUCCGGUGGUGCACCACCAUCACCAUGACACCCAUGCCGCCGUGGUG-CAUCACAGUAUUCCGGCUCAC------ ............(((.((.....))....((((((((((((......((((.....)))).....))))))-))))))........)))....------ ( -30.00, z-score = -1.99, R) >droYak2.chr3L 10991386 92 + 24197627 ACCACUACGCUCGUCUGCAGUCCGCUCCGGUGGUGCACCACCAUCACCAUGACACCCACGCCGCCGUGGUG-CAUCACAGUAUCCCAGCCCAC------ ........(((.(..(((((....))...((((((((((((........((.....)).......))))))-)))))).)))..).)))....------ ( -24.46, z-score = -0.27, R) >droEre2.scaffold_4784 10974000 92 + 25762168 ACCACUACGCUCGUCUGCAAGCCGCUCCGGUGGUGCACCACCACCACCAUGACACCCACGCCGCCGUGGUG-CAUCACAGUAUCCCAGCCCAC------ ........(((.(..(((..........(((((((......)))))))(((.((((.(((....)))))))-)))....)))..).)))....------ ( -26.50, z-score = -0.86, R) >droAna3.scaffold_13337 15879476 80 + 23293914 AUCACUAUGCCCACCUGCACGCCGCCCCCGUGCA---CCACUACACCACCCACUCGGCCAUCAAGACGGUGGUGCCAGCCCAC---------------- ........(((((((((((((.......))))))---........((........))..........))))).))........---------------- ( -18.70, z-score = -0.38, R) >dp4.chrXR_group6 7248021 77 - 13314419 ACCACUACGAGCAUCUGCAUGCCGCGCCGGUGGU---GCACCAUCACAGUCACG------CCGCUUUGGUG-CACUA-CAAGCCCCAU----------- .......((.((((....)))))).((..(((((---((((((....(((....------..))).)))))-)))))-)..)).....----------- ( -26.20, z-score = -1.27, R) >droPer1.super_47 27062 77 - 592741 ACCACUACGAGCAUCUGCAUGCCGCGCCGGUGGU---GCACCAUCACAGUCACG------CCGCUUUGGUG-CACUA-CAAGCCCCAU----------- .......((.((((....)))))).((..(((((---((((((....(((....------..))).)))))-)))))-)..)).....----------- ( -26.20, z-score = -1.27, R) >droWil1.scaffold_180916 60762 95 - 2700594 AUCAAGUGGCUCAUCUGCAUGCCGCUCCUGUGGU---CCAUCAUCACAGUCACAGUCACUCCGCCCUGGUG-CAUCAUCGUGCUGCCUAUCCUGCCCAU ....(((((((.....((.....))..(((((((---.....)))))))....)))))))..((...((((-(((....)))).)))......)).... ( -21.10, z-score = 0.51, R) >consensus ACCACUACGCUCGUCUGCACGCCGCUCCGGUGGUGCACCACCAUCACCAUGACACCCACGCCGCCGUGGUG_CAUCACAGUACCCCAGC_CAC______ ..(((((((.......((.....))...((((((.....))))))...................)))))))............................ (-11.85 = -12.23 + 0.39)

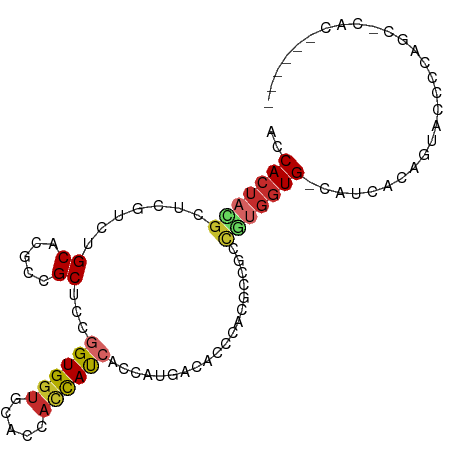
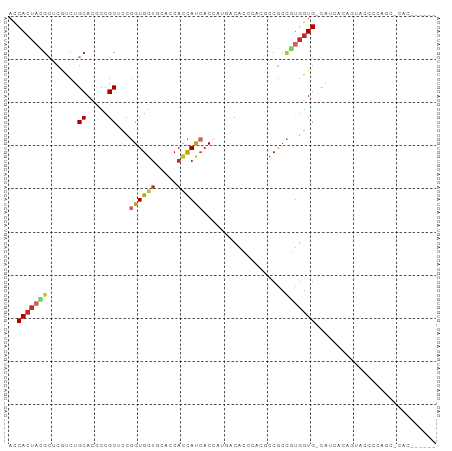
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:29 2011