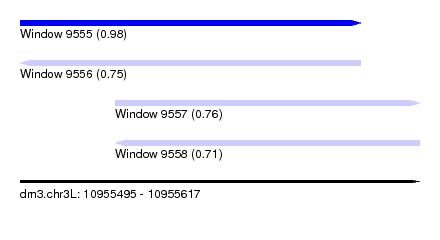
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,955,495 – 10,955,617 |
| Length | 122 |
| Max. P | 0.977244 |

| Location | 10,955,495 – 10,955,599 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Shannon entropy | 0.37583 |
| G+C content | 0.53602 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -24.81 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
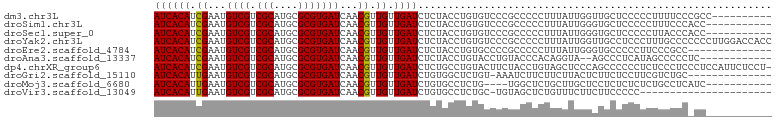
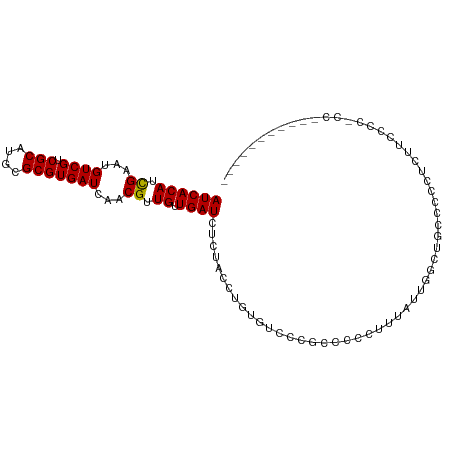
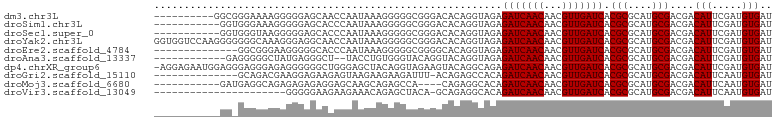
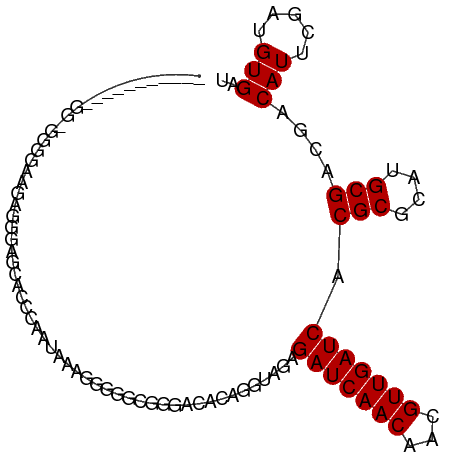
>dm3.chr3L 10955495 104 + 24543557 GCGCAUGCGC-----------GCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGUU (((((((((.-----------(((((..((..................))..))))))))))))))(.(((((((...))))))).).(((.(((............))).))). ( -32.47, z-score = -2.24, R) >droSim1.chr3L 10350244 104 + 22553184 GCGCAUGCGC-----------GCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGGU (((((((((.-----------(((((..((..................))..))))))))))))))(.(((((((...))))))).)...............(((......))). ( -35.57, z-score = -2.66, R) >droSec1.super_0 3179248 104 + 21120651 GCGCAUGCGC-----------GCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGGU (((((((((.-----------(((((..((..................))..))))))))))))))(.(((((((...))))))).)...............(((......))). ( -35.57, z-score = -2.66, R) >droYak2.chr3L 10979245 104 + 24197627 GCGCAUGCGC-----------GCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGUU (((((((((.-----------(((((..((..................))..))))))))))))))(.(((((((...))))))).).(((.(((............))).))). ( -32.47, z-score = -2.24, R) >droEre2.scaffold_4784 10963287 104 + 25762168 GCGCAUGCGC-----------GCGGCGCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGCCCCGCCCCCUUUAUUGGGU (((((((((.-----------((((((.((..................)).)))))))))))))))(.(((((((...))))))).)...............(((......))). ( -36.27, z-score = -1.99, R) >droAna3.scaffold_13337 15870082 100 + 23293914 GCGCAUGCGC-----------GCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUA-CCUGUACCCACAGGUA--- (((((((((.-----------(((((..((..................))..))))))))))))))(.(((((((...))))))).)......((-(((((....)))))))--- ( -39.07, z-score = -4.27, R) >dp4.chrXR_group6 7237384 107 - 13314419 GCGCAUGCGCCUUGCGGAGGAGCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUGCCUGUA-CUUCUACCUGUA------- (((((((((.......((((((.....))))))..((((..((.....))...)))))))))))))(.(((((((...))))))).)........-............------- ( -33.90, z-score = -1.06, R) >droWil1.scaffold_180916 40803 82 - 2700594 GCGCAUGAGU-----------GCGG--CUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUGGUCGGG-------------------- (((((((...-----------((((--(..(((...............))).)))))..)))))))..(((((((...)))))))..........-------------------- ( -25.16, z-score = 0.12, R) >droGri2.scaffold_15110 11580120 96 - 24565398 GCGCAUGCGC-----------GCGG--CAUUUCAAACGAAAUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUGGCUCUG---UAAAUCUUCUUCUU--- (((((((((.-----------((((--((((.(((...........))))))))))))))))))))..(((((((...)))))))..........---..............--- ( -34.60, z-score = -2.89, R) >droMoj3.scaffold_6680 3547944 96 - 24764193 GCGCAUGCGC-----------GCGG--CGCUUCAAACGAAAUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUGCCUCUG---UGGCUCUGCUUGCU--- (((((((((.-----------((((--((.(((((...........))))))))))))))))))))..(((((((...))))))).(.(((....---.))).)........--- ( -38.80, z-score = -2.24, R) >droVir3.scaffold_13049 15627521 99 + 25233164 GCGCAUGCGC-----------GCGG--CGCUUCAAACGAAAUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUGCCUCUGCUGUAGCUCUGUUUCUU--- (((((((((.-----------((((--((.(((((...........))))))))))))))))))))..(((((((...))))))).(.((..(....)..)).)........--- ( -35.70, z-score = -1.63, R) >consensus GCGCAUGCGC___________GCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUG_CCCGCCCCCUUUAUUG___ (((((((((..............((......))..((((..((.....))...)))))))))))))..(((((((...))))))).............................. (-24.81 = -24.90 + 0.09)

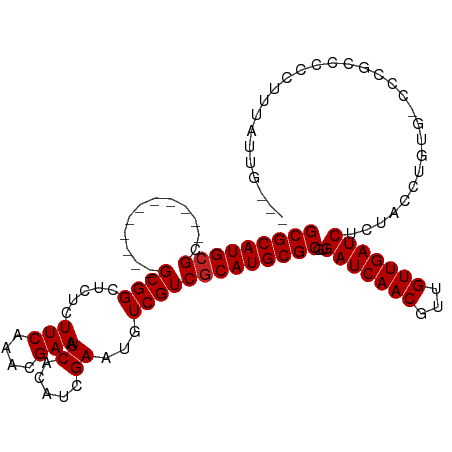
| Location | 10,955,495 – 10,955,599 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Shannon entropy | 0.37583 |
| G+C content | 0.53602 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.35 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10955495 104 - 24543557 AACCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGC-----------GCGCAUGCGC ..((......))..................(((((((...)))))))..(((((((((.((.....))..(((.((((.......))))..)))-----------.))))))))) ( -31.10, z-score = -0.75, R) >droSim1.chr3L 10350244 104 - 22553184 ACCCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGC-----------GCGCAUGCGC .(((......))).................(((((((...)))))))..(((((((((.((.....))..(((.((((.......))))..)))-----------.))))))))) ( -34.40, z-score = -1.54, R) >droSec1.super_0 3179248 104 - 21120651 ACCCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGC-----------GCGCAUGCGC .(((......))).................(((((((...)))))))..(((((((((.((.....))..(((.((((.......))))..)))-----------.))))))))) ( -34.40, z-score = -1.54, R) >droYak2.chr3L 10979245 104 - 24197627 AACCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGC-----------GCGCAUGCGC ..((......))..................(((((((...)))))))..(((((((((.((.....))..(((.((((.......))))..)))-----------.))))))))) ( -31.10, z-score = -0.75, R) >droEre2.scaffold_4784 10963287 104 - 25762168 ACCCAAUAAAGGGGGCGGGGCACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGCGCCGC-----------GCGCAUGCGC .(((......))).((((.((.........(((((((...)))))))...(((((.(((......))))))))..............)).))))-----------(((....))) ( -34.50, z-score = -0.59, R) >droAna3.scaffold_13337 15870082 100 - 23293914 ---UACCUGUGGGUACAGG-UACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGC-----------GCGCAUGCGC ---((((((((........-))))))))..(((((((...)))))))..(((((((((.((.....))..(((.((((.......))))..)))-----------.))))))))) ( -39.10, z-score = -3.04, R) >dp4.chrXR_group6 7237384 107 + 13314419 -------UACAGGUAGAAG-UACAGGCAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGCUCCUCCGCAAGGCGCAUGCGC -------............-..........(((((((...)))))))..(((((((((.........((.(((.((((.......))))..)))))..((....))))))))))) ( -32.90, z-score = -0.76, R) >droWil1.scaffold_180916 40803 82 + 2700594 --------------------CCCGACCACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAG--CCGC-----------ACUCAUGCGC --------------------..........(((((((...)))))))..(((((((.(.((.....)).((((..(((.......)))--.)))-----------)).))))))) ( -22.20, z-score = -0.79, R) >droGri2.scaffold_15110 11580120 96 + 24565398 ---AAGAAGAAGAUUUA---CAGAGCCACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAUUUCGUUUGAAAUG--CCGC-----------GCGCAUGCGC ---..............---..........(((((((...)))))))..(((((((((.((.(((((((((......)).))).))))--...)-----------)))))))))) ( -29.10, z-score = -1.67, R) >droMoj3.scaffold_6680 3547944 96 + 24764193 ---AGCAAGCAGAGCCA---CAGAGGCACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAUUUCGUUUGAAGCG--CCGC-----------GCGCAUGCGC ---..........(((.---....)))...(((((((...)))))))..(((((((((.((...(((((((......)).)))))...--...)-----------)))))))))) ( -33.40, z-score = -1.40, R) >droVir3.scaffold_13049 15627521 99 - 25233164 ---AAGAAACAGAGCUACAGCAGAGGCACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAUUUCGUUUGAAGCG--CCGC-----------GCGCAUGCGC ---..........(((........)))...(((((((...)))))))..(((((((((.((...(((((((......)).)))))...--...)-----------)))))))))) ( -31.20, z-score = -0.87, R) >consensus ___CAAUAAAGGGGGCAGG_CACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGC___________GCGCAUGCGC ..............................(((((((...)))))))..(((((((((....(((.....))).((((....))))....................))))))))) (-24.35 = -24.35 + 0.01)

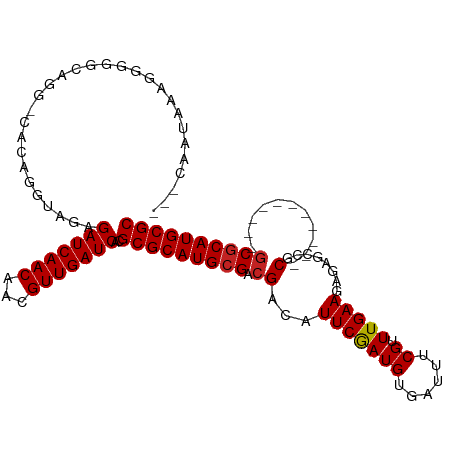
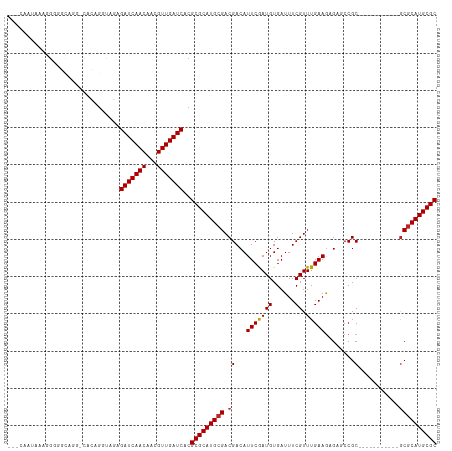
| Location | 10,955,524 – 10,955,617 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 72.00 |
| Shannon entropy | 0.58983 |
| G+C content | 0.53588 |
| Mean single sequence MFE | -17.81 |
| Consensus MFE | -9.70 |
| Energy contribution | -9.49 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10955524 93 + 24543557 AUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGUUGCUCCCCCUUUUCCCGCC---------- ....((((((......))).))).(((.(((((((...)))))))..............((..((......))..))............))).---------- ( -15.30, z-score = -0.45, R) >droSim1.chr3L 10350273 92 + 22553184 AUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGGUGCUCCCCCUUUCCCACC----------- ..((((.((.((((....)))).)).(.(((((((...))))))).)....))))....((.(((......))).))...............----------- ( -19.50, z-score = -1.33, R) >droSec1.super_0 3179277 92 + 21120651 AUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGGUGCUCCCCCUUACCCACC----------- ..((((.((.((((....)))).)).(.(((((((...))))))).)....))))....((.(((......))).))...............----------- ( -19.50, z-score = -1.14, R) >droYak2.chr3L 10979274 103 + 24197627 AUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGUUGCCUCCCUUUGCCCCCCCUUGGACCACC ((((((.((...((((.(((....)))))))...)).)).)))).......((.((((.....((......))..((........))........)))))).. ( -16.00, z-score = 0.62, R) >droEre2.scaffold_4784 10963316 89 + 25762168 AUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGCCCCGCCCCCUUUAUUGGGUGCCCCCUUCCCGCC-------------- ....((((((......))).))).(((.(((((((...)))))))..............((.(((......))).))........))).-------------- ( -19.40, z-score = -0.74, R) >droAna3.scaffold_13337 15870111 89 + 23293914 AUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUACCUGUACCCACAGGUA--AGCCCUCAUAGCCCCCUC------------ ....((((((......))).))).(((.(((((((...))))))).)......(((((((....)))))))--.))...............------------ ( -19.80, z-score = -1.32, R) >dp4.chrXR_group6 7237424 102 - 13314419 AUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUGCCUGUACUUCUACCUGUAGCUCCCAGCCCCCCUCUCCCUCCCUCCAUUCUCCU- ........(((((..((.(((.(((.(.(((((((...))))))).)))).)))))...........((.....)).................)))))....- ( -18.00, z-score = -2.62, R) >droGri2.scaffold_15110 11580147 88 - 24565398 AUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUGGCUCUGU-AAAUCUUCUUCUUACUCUUCUCCUUCGUCUGC-------------- .(((((...........(((....))).(((((((...))))))))))))......-................................-------------- ( -12.80, z-score = 0.48, R) >droMoj3.scaffold_6680 3547971 88 - 24764193 AUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUGCCUCUG----UGGCUCUGCUUGCUCCUCUCUCUCUGCCUCAUC----------- .......(((.....((((((.(((((.(((((((...))))))))))))...))----)))).......((............)).)))..----------- ( -20.60, z-score = -1.49, R) >droVir3.scaffold_13049 15627548 80 + 25233164 AUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUGCCUCUGC-UGUAGCUCUGUUUCUUCUUCCCCC---------------------- ............((..(.(((.(((((.(((((((...))))))))))))...)))-.)..))..................---------------------- ( -17.20, z-score = -1.43, R) >consensus AUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUACCUGUGUCCCGCCCCCUUUAUUGGCUGCCCCCUCUUCCCC_CC___________ ((((((.((...((((.(((....)))))))...)).)).))))........................................................... ( -9.70 = -9.49 + -0.21)
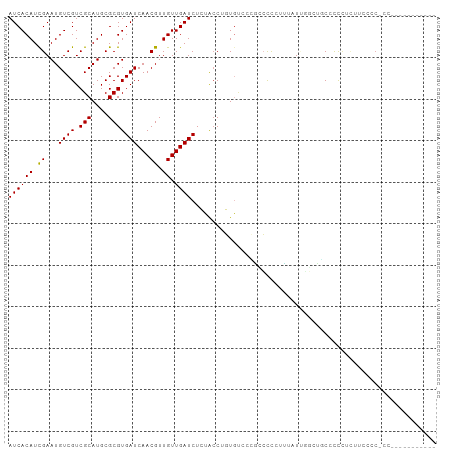
| Location | 10,955,524 – 10,955,617 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 72.00 |
| Shannon entropy | 0.58983 |
| G+C content | 0.53588 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -10.50 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10955524 93 - 24543557 ----------GGCGGGAAAAGGGGGAGCAACCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAU ----------.(((............((..((......))..))..............(((((((...))))))).)))((((.(((......)))))))... ( -22.70, z-score = -1.54, R) >droSim1.chr3L 10350273 92 - 22553184 -----------GGUGGGAAAGGGGGAGCACCCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAU -----------.....(((.(..(..((((((......))).(((.(((....))...(((((((...))))))).).))).)))..)..).)))........ ( -24.30, z-score = -1.29, R) >droSec1.super_0 3179277 92 - 21120651 -----------GGUGGGUAAGGGGGAGCACCCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAU -----------.((.((...(..(..((((((......))).(((.(((....))...(((((((...))))))).).))).)))..)..)..)).))..... ( -22.60, z-score = -0.43, R) >droYak2.chr3L 10979274 103 - 24197627 GGUGGUCCAAGGGGGGGCAAAGGGAGGCAACCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAU .(((((((......))))........((..((......))..))....))).......(((((((...)))))))...(((((.(((......)))))))).. ( -28.20, z-score = -1.62, R) >droEre2.scaffold_4784 10963316 89 - 25762168 --------------GGCGGGAAGGGGGCACCCAAUAAAGGGGGCGGGGCACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAU --------------.(((........((.(((......))).))..............(((((((...))))))).)))((((.(((......)))))))... ( -23.80, z-score = -0.81, R) >droAna3.scaffold_13337 15870111 89 - 23293914 ------------GAGGGGGCUAUGAGGGCU--UACCUGUGGGUACAGGUACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAU ------------.....((((.....))))--((((((((........))))))))..(((((((...)))))))...(((((.(((......)))))))).. ( -25.90, z-score = -1.13, R) >dp4.chrXR_group6 7237424 102 + 13314419 -AGGAGAAUGGAGGGAGGGAGAGGGGGGCUGGGAGCUACAGGUAGAAGUACAGGCAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAU -....(((((..(.............(.(((....(((....))).....))).)...(((((((...))))))).(((....))).)..)))))........ ( -23.70, z-score = -1.64, R) >droGri2.scaffold_15110 11580147 88 + 24565398 --------------GCAGACGAAGGAGAAGAGUAAGAAGAAGAUUU-ACAGAGCCACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAU --------------((...(....)......(((((.......)))-))...))(((((((((((...))))))).(((....)))...........)))).. ( -16.40, z-score = -1.07, R) >droMoj3.scaffold_6680 3547971 88 + 24764193 -----------GAUGAGGCAGAGAGAGAGGAGCAAGCAGAGCCA----CAGAGGCACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAU -----------......(((..(((.(.(..(((.((...(((.----....)))...(((((((...)))))))....)).)))..)..).)))..)))... ( -23.80, z-score = -2.48, R) >droVir3.scaffold_13049 15627548 80 - 25233164 ----------------------GGGGGAAGAAGAAACAGAGCUACA-GCAGAGGCACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAU ----------------------..................((....-)).....(((((((((((...))))))).(((....)))...........)))).. ( -15.30, z-score = -0.44, R) >consensus ___________GG_GGGGAAGAGGGAGCACCCAAUAAAGGGGGCGGGACACAGGUAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAU ..........................................................(((((((...))))))).(((....)))....(((.....))).. (-10.50 = -10.50 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:28 2011