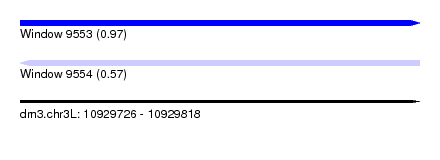
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,929,726 – 10,929,818 |
| Length | 92 |
| Max. P | 0.972878 |

| Location | 10,929,726 – 10,929,818 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.98 |
| Shannon entropy | 0.30644 |
| G+C content | 0.49288 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
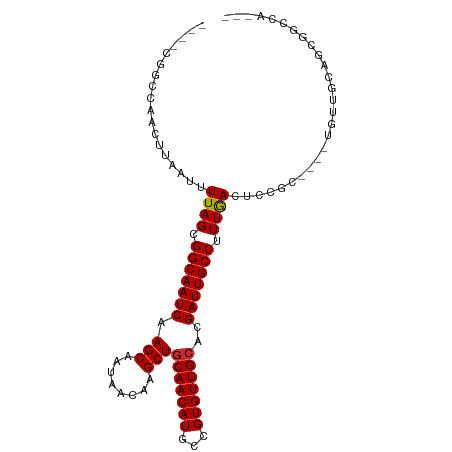
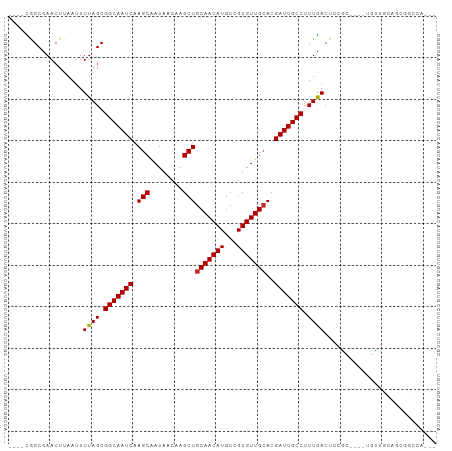
>dm3.chr3L 10929726 92 + 24543557 ----CGGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCCGC----UGUUACAGCGGCCA--- ----.((((.........((((.(((((((.(((........)))(((((((...)))))))..))))))).))))....((----((...)))))))).--- ( -35.40, z-score = -3.72, R) >droSim1.chr3L 10324377 92 + 22553184 ----CGGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCCGC----UGUUGCAGCGGCCA--- ----.((((.........((((.(((((((.(((........)))(((((((...)))))))..))))))).))))....((----((...)))))))).--- ( -35.40, z-score = -2.99, R) >droSec1.super_0 3154021 92 + 21120651 ----CGGCCAACUUAAUUUUAGCGGCAAUCAAGCGAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCCGC----UGUUGCAGCGGCCA--- ----.((((.........((((.(((((((.(((........)))(((((((...)))))))..))))))).))))....((----((...)))))))).--- ( -36.10, z-score = -3.05, R) >droEre2.scaffold_4784 10936272 92 + 25762168 ----CAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCCGC----UGCAACAUCGGGCA--- ----..(((..........(((((((((((.(((........)))(((((((...)))))))..)))))))...(....)))----))........))).--- ( -27.97, z-score = -1.58, R) >droAna3.scaffold_13337 15840323 82 + 23293914 ----CAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGGACGAUUGCCUUUGACUCCGC----AGCA------------- ----.................(((((((((.(((........))).((((((...))))))...)))))))..((......)----))).------------- ( -19.10, z-score = 0.01, R) >dp4.chrXR_group6 7210253 83 - 13314419 ----CGGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCUGA----GUCCG------------ ----(((...(((((...((((.(((((((.(((........)))(((((((...)))))))..))))))).))))...)))----)))))------------ ( -27.10, z-score = -3.00, R) >droPer1.super_2310 5150 83 - 5321 ----CGGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCUGA----GUCCG------------ ----(((...(((((...((((.(((((((.(((........)))(((((((...)))))))..))))))).))))...)))----)))))------------ ( -27.10, z-score = -3.00, R) >droVir3.scaffold_13049 15598534 99 + 25233164 CCAAUAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUAACUCGAC----UCUGGCCGUGGCUCGUU (((...((((........((((.(((((((.(((........)))(((((((...)))))))..))))))).))))......----..))))..)))...... ( -30.09, z-score = -2.29, R) >droGri2.scaffold_15110 11557965 100 - 24565398 CCAAUAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCGAAGUGGCUUGGCCGCGGCUC--- .....((((.........((((.(((((((.(((........)))(((((((...)))))))..))))))).))))......(((((...))))))))).--- ( -34.90, z-score = -2.47, R) >consensus ____CGGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCCGC____UGUUGCAGCGGCCA___ ..................((((.(((((((.(((........)))(((((((...)))))))..))))))).))))........................... (-21.53 = -21.54 + 0.01)

| Location | 10,929,726 – 10,929,818 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.98 |
| Shannon entropy | 0.30644 |
| G+C content | 0.49288 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -23.49 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10929726 92 - 24543557 ---UGGCCGCUGUAACA----GCGGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCCG---- ---.(((((((.....(----((((((((((..((((.(((((......))))))))).((........)))))))).))))).......)).))))).---- ( -34.90, z-score = -2.10, R) >droSim1.chr3L 10324377 92 - 22553184 ---UGGCCGCUGCAACA----GCGGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCCG---- ---.(((((((((((((----((...((.....))...((((...)))))).)))))))))........((((((((........)))))))).)))).---- ( -35.70, z-score = -1.99, R) >droSec1.super_0 3154021 92 - 21120651 ---UGGCCGCUGCAACA----GCGGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUCGCUUGAUUGCCGCUAAAAUUAAGUUGGCCG---- ---.(((((((((((((----((...((.....))...((((...)))))).)))))))))........((((((((........)))))))).)))).---- ( -35.80, z-score = -2.06, R) >droEre2.scaffold_4784 10936272 92 - 25762168 ---UGCCCGAUGUUGCA----GCGGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUG---- ---.(((....(((..(----((((((((((..((((.(((((......))))))))).((........)))))))).)))))..)))......)))..---- ( -30.00, z-score = -0.65, R) >droAna3.scaffold_13337 15840323 82 - 23293914 -------------UGCU----GCGGAGUCAAAGGCAAUCGUCCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUG---- -------------....----((((((((((..((((.(((((.....)).))))))).((........)))))))).)))).................---- ( -20.40, z-score = 0.79, R) >dp4.chrXR_group6 7210253 83 + 13314419 ------------CGGAC----UCAGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCCG---- ------------..(((----.....)))...(((((((((((((((.....)))))))((........)).))))))))(((((.......)))))..---- ( -26.50, z-score = -1.55, R) >droPer1.super_2310 5150 83 + 5321 ------------CGGAC----UCAGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCCG---- ------------..(((----.....)))...(((((((((((((((.....)))))))((........)).))))))))(((((.......)))))..---- ( -26.50, z-score = -1.55, R) >droVir3.scaffold_13049 15598534 99 - 25233164 AACGAGCCACGGCCAGA----GUCGAGUUAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUAUUGG ......(((.((((((.----......(((..(((((((((((((((.....)))))))((........)).))))))))..))).......))))))..))) ( -31.94, z-score = -1.37, R) >droGri2.scaffold_15110 11557965 100 + 24565398 ---GAGCCGCGGCCAAGCCACUUCGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUAUUGG ---.((((((((((((((.....(((((.....((((.(((((......))))))))).))))).....)))))...)))))...(((...)))))))..... ( -33.40, z-score = -1.06, R) >consensus ___UGGCCGCUGCAACA____GCGGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCCG____ ................................(((((((((((((((.....)))))))((........)).))))))))(((((.......)))))...... (-23.49 = -23.60 + 0.11)
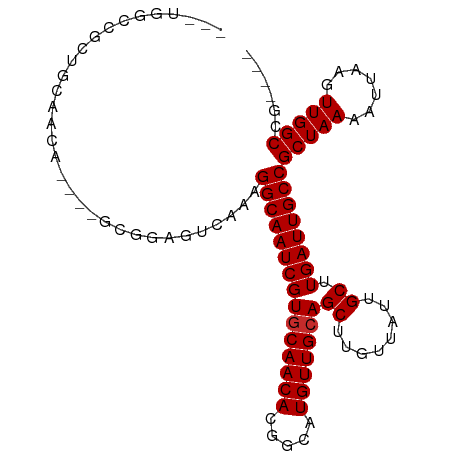
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:25 2011