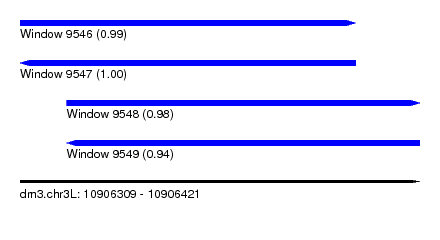
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,906,309 – 10,906,421 |
| Length | 112 |
| Max. P | 0.999055 |

| Location | 10,906,309 – 10,906,403 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.28 |
| Shannon entropy | 0.41833 |
| G+C content | 0.58690 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -26.90 |
| Energy contribution | -26.06 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
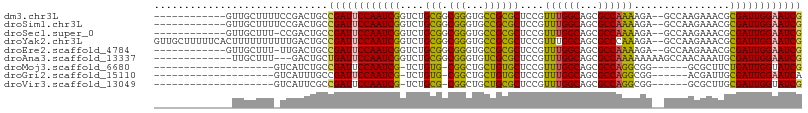
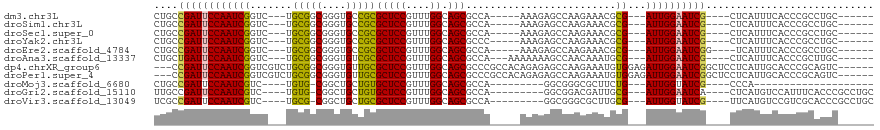
>dm3.chr3L 10906309 94 + 24543557 ------------GUUGCUUUUCCGACUGCCGAUUCCAAUCGGUCUGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCAAAAGA--GCCAAGAAACGCGAUUGGAAUCG ------------((((......))))...((((((((((((.((((((((....)))))((((..((((((...)))))).))--))..)))....)))))))))))) ( -41.80, z-score = -2.75, R) >droSim1.chr3L 10300603 94 + 22553184 ------------GUUGCUUUUCCGACUGCCGAUUCCAAUCGGUCUGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCAAAAGA--GCCAAGAAACGCGAUUGGAAUCG ------------((((......))))...((((((((((((.((((((((....)))))((((..((((((...)))))).))--))..)))....)))))))))))) ( -41.80, z-score = -2.75, R) >droSec1.super_0 3129836 93 + 21120651 ------------GUUGCUUU-CCGACUGCCGAUUCCAAUCGGUCUGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCAAAAGA--GCCAAGAAACGCGAUUGGAAUCG ------------((((....-.))))...((((((((((((.((((((((....)))))((((..((((((...)))))).))--))..)))....)))))))))))) ( -41.60, z-score = -2.73, R) >droYak2.chr3L 10926627 106 + 24197627 GUUGCUUUUUCACUUUUUUUUUUGACUGCCGAUUCCAAUCGGUCUGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCCAAAGA--GCCAAGAAACGCGAUUGGAAUCG .............................((((((((((((.(((..(((((((((.((.(.......))).)))))).....--))).)))....)))))))))))) ( -37.10, z-score = -1.30, R) >droEre2.scaffold_4784 10912721 93 + 25762168 ------------GUUGCUUU-UUGACUGCCGAUUCCAAUCGGUCUGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCAAAAGA--GCCAAGAAACGCGAUUGGAAUCG ------------........-........((((((((((((.((((((((....)))))((((..((((((...)))))).))--))..)))....)))))))))))) ( -40.40, z-score = -2.59, R) >droAna3.scaffold_13337 15814189 92 + 23293914 -------------UUGCUUU---GACUGCUGAUUCCAAUCGGUCUGCGGCGGGUGUCGCGCUCCGUUUGGCAGCGCCAAAAAAAAGCCAACAAAUGCGAUUGGAAUCG -------------..((...---....)).(((((((((((....(((((....)))))(((...((((((...))))))....))).........))))))))))). ( -33.80, z-score = -1.89, R) >droMoj3.scaffold_6680 3491922 80 - 24764193 --------------------GUCAUCUGCCGAUUCCAAUCG-UCUGUG-CGGCUGCUGUGCUCCGUUUGGCAGCGCCAGGCGG------GCGCUUCUGAUUGGUAUCG --------------------.........((((.(((((((-...(((-((((......)))(((((((((...)))))))))------))))...))))))).)))) ( -34.50, z-score = -2.45, R) >droGri2.scaffold_15110 11536559 80 - 24565398 --------------------GUCAUUUGCCGAUUCCAAUCG-UCUGUG-CGGCUGCUGUGCUCCGUUUGGCAGCGCCAGGCGG------ACGAUUGCGAUUGGAAUCA --------------------..........(((((((((((-(..(..-(((...)))..)((((((((((...)))))))))------).....)))))))))))). ( -37.50, z-score = -3.33, R) >droVir3.scaffold_13049 15573000 80 + 25233164 --------------------GUCAUUCGCCGAUUCCAAUCG-UCUGCG-CGGCUGCUGCGCUCCGUUUGGCAGCGCCAGGCGG------GCGCUUGCGAUUGGUAUCG --------------------.........((((.(((((((-(..(((-(.((....))...(((((((((...)))))))))------))))..)))))))).)))) ( -40.50, z-score = -2.95, R) >consensus _____________UUGCUUU_UCGACUGCCGAUUCCAAUCGGUCUGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCAAAAGA__GCCAAGAAACGCGAUUGGAAUCG .............................((((((((((((....(((.(((...))))))....((((((...))))))................)))))))))))) (-26.90 = -26.06 + -0.85)
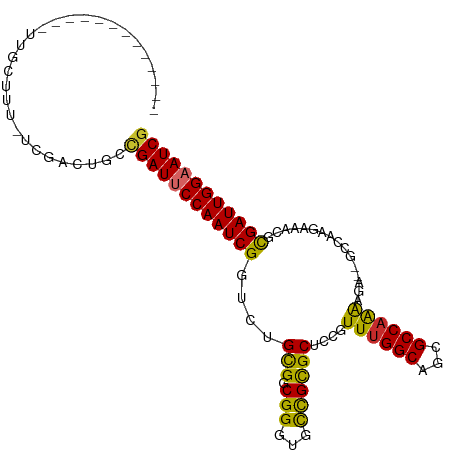
| Location | 10,906,309 – 10,906,403 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.28 |
| Shannon entropy | 0.41833 |
| G+C content | 0.58690 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -24.56 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10906309 94 - 24543557 CGAUUCCAAUCGCGUUUCUUGGC--UCUUUUGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCAGACCGAUUGGAAUCGGCAGUCGGAAAAGCAAC------------ ((((((((((((.((((....((--(((((((((...)))))).)))))(((((....)))))))))))))))))))))((..........))...------------ ( -45.50, z-score = -4.14, R) >droSim1.chr3L 10300603 94 - 22553184 CGAUUCCAAUCGCGUUUCUUGGC--UCUUUUGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCAGACCGAUUGGAAUCGGCAGUCGGAAAAGCAAC------------ ((((((((((((.((((....((--(((((((((...)))))).)))))(((((....)))))))))))))))))))))((..........))...------------ ( -45.50, z-score = -4.14, R) >droSec1.super_0 3129836 93 - 21120651 CGAUUCCAAUCGCGUUUCUUGGC--UCUUUUGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCAGACCGAUUGGAAUCGGCAGUCGG-AAAGCAAC------------ ((((((((((((.((((....((--(((((((((...)))))).)))))(((((....)))))))))))))))))))))((......-...))...------------ ( -45.60, z-score = -4.19, R) >droYak2.chr3L 10926627 106 - 24197627 CGAUUCCAAUCGCGUUUCUUGGC--UCUUUGGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCAGACCGAUUGGAAUCGGCAGUCAAAAAAAAAAGUGAAAAAGCAAC ((((((((((((.((((....((--(((...(((...)))....)))))(((((....)))))))))))))))))))))((..(((..........)))....))... ( -43.20, z-score = -3.38, R) >droEre2.scaffold_4784 10912721 93 - 25762168 CGAUUCCAAUCGCGUUUCUUGGC--UCUUUUGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCAGACCGAUUGGAAUCGGCAGUCAA-AAAGCAAC------------ ((((((((((((.((((....((--(((((((((...)))))).)))))(((((....)))))))))))))))))))))((......-...))...------------ ( -45.60, z-score = -4.95, R) >droAna3.scaffold_13337 15814189 92 - 23293914 CGAUUCCAAUCGCAUUUGUUGGCUUUUUUUUGGCGCUGCCAAACGGAGCGCGACACCCGCCGCAGACCGAUUGGAAUCAGCAGUC---AAAGCAA------------- .(((((((((((..(((((.(((.......(.(((((.((....))))))).).....)))))))).))))))))))).((....---...))..------------- ( -35.40, z-score = -3.00, R) >droMoj3.scaffold_6680 3491922 80 + 24764193 CGAUACCAAUCAGAAGCGC------CCGCCUGGCGCUGCCAAACGGAGCACAGCAGCCG-CACAGA-CGAUUGGAAUCGGCAGAUGAC-------------------- ((((.((((((....(.((------(((..((((...))))..))).)).).((....)-).....-.)))))).)))).........-------------------- ( -26.80, z-score = -1.99, R) >droGri2.scaffold_15110 11536559 80 + 24565398 UGAUUCCAAUCGCAAUCGU------CCGCCUGGCGCUGCCAAACGGAGCACAGCAGCCG-CACAGA-CGAUUGGAAUCGGCAAAUGAC-------------------- .((((((((((((.....(------(((..((((...))))..)))).....((....)-)...).-)))))))))))..........-------------------- ( -28.80, z-score = -2.26, R) >droVir3.scaffold_13049 15573000 80 - 25233164 CGAUACCAAUCGCAAGCGC------CCGCCUGGCGCUGCCAAACGGAGCGCAGCAGCCG-CGCAGA-CGAUUGGAAUCGGCGAAUGAC-------------------- ((((.((((((((..((((------(((..((((...))))..))).)))).((.....-.)).).-))))))).)))).........-------------------- ( -34.30, z-score = -2.38, R) >consensus CGAUUCCAAUCGCGUUUCUUGGC__UCUUUUGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCAGACCGAUUGGAAUCGGCAGUCGA_AAAGCAA_____________ ((((((((((((....................(((((.((....))))))).((.......))....))))))))))))............................. (-24.56 = -25.48 + 0.91)

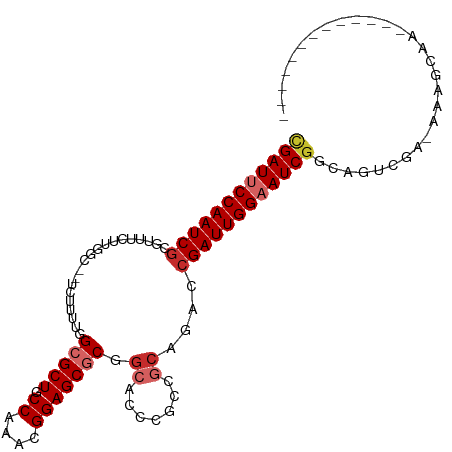
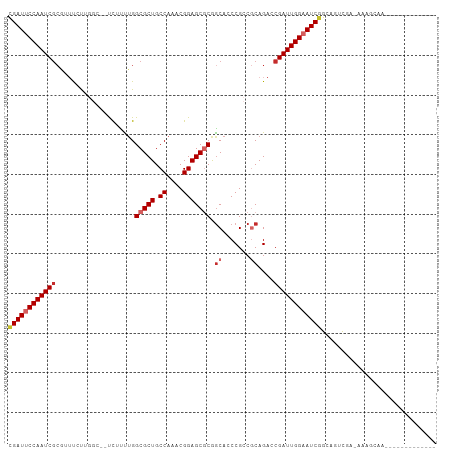
| Location | 10,906,322 – 10,906,421 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Shannon entropy | 0.47268 |
| G+C content | 0.61418 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -21.86 |
| Energy contribution | -21.63 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10906322 99 + 24543557 CUGCCGAUUCCAAUCGGUC---UGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCA-----AAAGAGCCAAGAAACGCG---AUUGGAAUCG----CUCAUUUCACCCGCCUGC------ ..((((((....)))))).---.((((((((((..(.((((..((((((...))))-----)).))))).......(((---((....))))----)......)))))))).))------ ( -41.50, z-score = -2.05, R) >droSim1.chr3L 10300616 99 + 22553184 CUGCCGAUUCCAAUCGGUC---UGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCA-----AAAGAGCCAAGAAACGCG---AUUGGAAUCG----CUCAUUUCACCCGCCUGC------ ..((((((....)))))).---.((((((((((..(.((((..((((((...))))-----)).))))).......(((---((....))))----)......)))))))).))------ ( -41.50, z-score = -2.05, R) >droSec1.super_0 3129848 99 + 21120651 CUGCCGAUUCCAAUCGGUC---UGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCA-----AAAGAGCCAAGAAACGCG---AUUGGAAUCG----CUCAUUUCACCCGCCUGC------ ..((((((....)))))).---.((((((((((..(.((((..((((((...))))-----)).))))).......(((---((....))))----)......)))))))).))------ ( -41.50, z-score = -2.05, R) >droYak2.chr3L 10926652 99 + 24197627 CUGCCGAUUCCAAUCGGUC---UGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCC-----AAAGAGCCAAGAAACGCG---AUUGGAAUCG----CUCAUUUCACCCGCCUGC------ ..((((((....)))))).---.((((((((((..(((((((....)).)))))..-----...............(((---((....))))----)......)))))))).))------ ( -39.40, z-score = -1.34, R) >droEre2.scaffold_4784 10912733 99 + 25762168 CUGCCGAUUCCAAUCGGUC---UGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCA-----AAAGAGCCAAGAAACGCG---AUUGGAAUCGG----UCAUUUCACCCGCCUGC------ ..((((((((((((((.((---((((((....)))))((((..((((((...))))-----)).))))..)))....))---)))))))))))----)................------ ( -45.70, z-score = -3.10, R) >droAna3.scaffold_13337 15814198 101 + 23293914 CUGCUGAUUCCAAUCGGUC---UGCGGCGGGUGUCGCGCUCCGUUUGGCAGCGCCA---AAAAAAAGCCAACAAAUGCG---AUUGGAAUCG----CUCAUUUCACCCGCUUGC------ ..((((((....)))))).---.((((((((((..(((((((....)).)))))..---.................(((---((....))))----)......))))))).)))------ ( -34.90, z-score = -1.20, R) >dp4.chrXR_group6 7179816 111 - 13314419 ---CCGAUUCCAAUCGGUCGUCUGCGGCGGGUGUUGCGCUCCGUUUGGCAGCGCCCGCCACAGAGAGCCAAGAAAUGUGGAGAUUGGAAUCGGCUCCUCAUUGCACCCGCAGUC------ ---(((((((((((((((..((((.((((((((((((.(.......))))))))))))).))))..))).(....).....))))))))))))......(((((....))))).------ ( -58.30, z-score = -5.30, R) >droPer1.super_4 3582708 111 + 7162766 ---CCGAUUCCAAUCGGUCGUCUGCGGCGGGUGUUGCGCUCCGUUUGGCAGCGCCCGCCACAGAGAGCCAAGAAAUGUGGAGAUUGGAAUCGGCUCCUCAUUGCACCCGCAGUC------ ---(((((((((((((((..((((.((((((((((((.(.......))))))))))))).))))..))).(....).....))))))))))))......(((((....))))).------ ( -58.30, z-score = -5.30, R) >droMoj3.scaffold_6680 3491927 79 - 24764193 CUGCCGAUUCCAAUCGUC----UGUG-CGGCUGCUGUGCUCCGUUUGGCAGCGCCA---------GGCGGGCGCUUCUG---AUUGGUAUCG----CCCA-------------------- ....((((.(((((((..----.(((-((((......)))(((((((((...))))---------)))))))))...))---))))).))))----....-------------------- ( -34.90, z-score = -1.84, R) >droGri2.scaffold_15110 11536564 99 - 24565398 UUGCCGAUUCCAAUCGUC----UGUG-CGGCUGCUGUGCUCCGUUUGGCAGCGCCA---------GGCGGACGAUUGCG---AUUGGAAUCA----CUCAUGUCCAUUUCACCCGCCUGC .....((((((((((((.----.(..-(((...)))..)((((((((((...))))---------)))))).....)))---))))))))).----........................ ( -37.50, z-score = -1.72, R) >droVir3.scaffold_13049 15573005 99 + 25233164 UCGCCGAUUCCAAUCGUC----UGCG-CGGCUGCUGCGCUCCGUUUGGCAGCGCCA---------GGCGGGCGCUUGCG---AUUGGUAUCG----UUCAUGUCCGUCGCACCCGCCUGC ..((.(((.......)))----.))(-(((.(((.(((((((....)).)))))..---------((((((((...(((---((....))))----)...))))))))))).)))).... ( -41.90, z-score = -0.59, R) >consensus CUGCCGAUUCCAAUCGGUC___UGCGGCGGGUGCCGCGCUCCGUUUGGCAGCGCCA_____AAAGAGCCAAGAAAUGCG___AUUGGAAUCG____CUCAUUUCACCCGCCUGC______ ....((((((((((........((((((....)))(((((((....)).)))))......................)))...))))))))))............................ (-21.86 = -21.63 + -0.23)
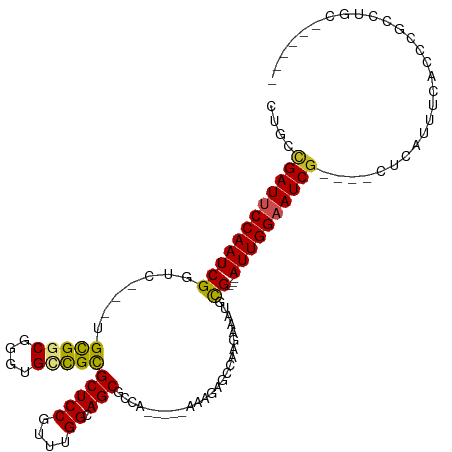
| Location | 10,906,322 – 10,906,421 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Shannon entropy | 0.47268 |
| G+C content | 0.61418 |
| Mean single sequence MFE | -43.35 |
| Consensus MFE | -19.61 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10906322 99 - 24543557 ------GCAGGCGGGUGAAAUGAG----CGAUUCCAAU---CGCGUUUCUUGGCUCUUU-----UGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCA---GACCGAUUGGAAUCGGCAG ------((..((...........)----)(((((((((---((.((((....(((((((-----((((...)))))).)))))(((((....))))))---)))))))))))))).)).. ( -46.60, z-score = -2.79, R) >droSim1.chr3L 10300616 99 - 22553184 ------GCAGGCGGGUGAAAUGAG----CGAUUCCAAU---CGCGUUUCUUGGCUCUUU-----UGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCA---GACCGAUUGGAAUCGGCAG ------((..((...........)----)(((((((((---((.((((....(((((((-----((((...)))))).)))))(((((....))))))---)))))))))))))).)).. ( -46.60, z-score = -2.79, R) >droSec1.super_0 3129848 99 - 21120651 ------GCAGGCGGGUGAAAUGAG----CGAUUCCAAU---CGCGUUUCUUGGCUCUUU-----UGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCA---GACCGAUUGGAAUCGGCAG ------((..((...........)----)(((((((((---((.((((....(((((((-----((((...)))))).)))))(((((....))))))---)))))))))))))).)).. ( -46.60, z-score = -2.79, R) >droYak2.chr3L 10926652 99 - 24197627 ------GCAGGCGGGUGAAAUGAG----CGAUUCCAAU---CGCGUUUCUUGGCUCUUU-----GGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCA---GACCGAUUGGAAUCGGCAG ------((..((...........)----)(((((((((---((.((((....(((((..-----.(((...)))....)))))(((((....))))))---)))))))))))))).)).. ( -43.60, z-score = -1.68, R) >droEre2.scaffold_4784 10912733 99 - 25762168 ------GCAGGCGGGUGAAAUGA----CCGAUUCCAAU---CGCGUUUCUUGGCUCUUU-----UGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCA---GACCGAUUGGAAUCGGCAG ------..............((.----(((((((((((---((.((((....(((((((-----((((...)))))).)))))(((((....))))))---)))))))))))))))))). ( -49.70, z-score = -3.98, R) >droAna3.scaffold_13337 15814198 101 - 23293914 ------GCAAGCGGGUGAAAUGAG----CGAUUCCAAU---CGCAUUUGUUGGCUUUUUUU---UGGCGCUGCCAAACGGAGCGCGACACCCGCCGCA---GACCGAUUGGAAUCAGCAG ------((..((...........)----)(((((((((---((..(((((.(((.......---(.(((((.((....))))))).).....))))))---)).))))))))))).)).. ( -37.80, z-score = -1.78, R) >dp4.chrXR_group6 7179816 111 + 13314419 ------GACUGCGGGUGCAAUGAGGAGCCGAUUCCAAUCUCCACAUUUCUUGGCUCUCUGUGGCGGGCGCUGCCAAACGGAGCGCAACACCCGCCGCAGACGACCGAUUGGAAUCGG--- ------...(((....)))........((((((((((((.(((.......))).(((((((((((((((((.((....))))))).....)))))))))).))..))))))))))))--- ( -49.80, z-score = -3.43, R) >droPer1.super_4 3582708 111 - 7162766 ------GACUGCGGGUGCAAUGAGGAGCCGAUUCCAAUCUCCACAUUUCUUGGCUCUCUGUGGCGGGCGCUGCCAAACGGAGCGCAACACCCGCCGCAGACGACCGAUUGGAAUCGG--- ------...(((....)))........((((((((((((.(((.......))).(((((((((((((((((.((....))))))).....)))))))))).))..))))))))))))--- ( -49.80, z-score = -3.43, R) >droMoj3.scaffold_6680 3491927 79 + 24764193 --------------------UGGG----CGAUACCAAU---CAGAAGCGCCCGCC---------UGGCGCUGCCAAACGGAGCACAGCAGCCG-CACA----GACGAUUGGAAUCGGCAG --------------------..(.----((((.(((((---(....(.(((((..---------((((...))))..))).)).).((....)-)...----...)))))).)))).).. ( -29.30, z-score = -1.56, R) >droGri2.scaffold_15110 11536564 99 + 24565398 GCAGGCGGGUGAAAUGGACAUGAG----UGAUUCCAAU---CGCAAUCGUCCGCC---------UGGCGCUGCCAAACGGAGCACAGCAGCCG-CACA----GACGAUUGGAAUCGGCAA ((..((..(((.......)))..)----)(((((((((---(((.....((((..---------((((...))))..)))).....((....)-)...----).))))))))))).)).. ( -32.40, z-score = 0.01, R) >droVir3.scaffold_13049 15573005 99 - 25233164 GCAGGCGGGUGCGACGGACAUGAA----CGAUACCAAU---CGCAAGCGCCCGCC---------UGGCGCUGCCAAACGGAGCGCAGCAGCCG-CGCA----GACGAUUGGAAUCGGCGA .(((((((((((..((....))..----((((....))---))...)))))))))---------)).(((((((((.((..((((.......)-))).----..)).))))...))))). ( -44.60, z-score = -1.82, R) >consensus ______GCAGGCGGGUGAAAUGAG____CGAUUCCAAU___CGCAUUUCUUGGCUCUUU_____UGGCGCUGCCAAACGGAGCGCGGCACCCGCCGCA___GACCGAUUGGAAUCGGCAG ............................((((((((((............................(((((.((....))))))).........((........)))))))))))).... (-19.61 = -20.17 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:21 2011