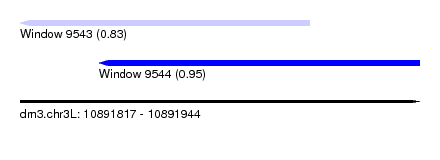
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,891,817 – 10,891,944 |
| Length | 127 |
| Max. P | 0.952962 |

| Location | 10,891,817 – 10,891,909 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 87.53 |
| Shannon entropy | 0.20676 |
| G+C content | 0.41464 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.06 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
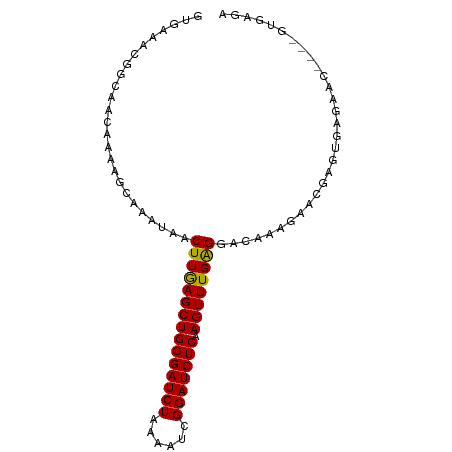
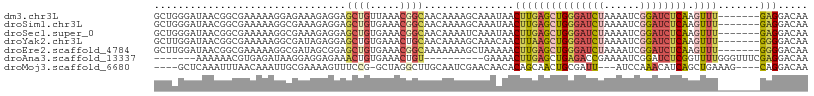
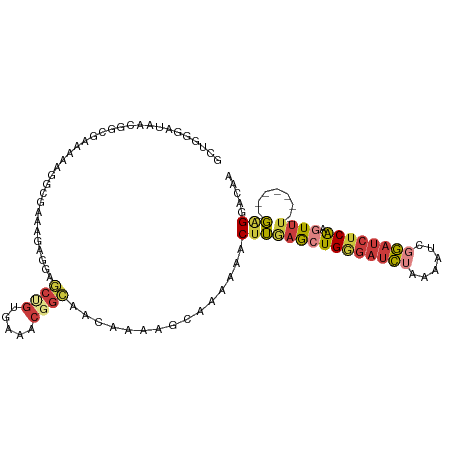
>dm3.chr3L 10891817 92 - 24543557 GUUAAACGGCAACAAAAGCAAAUAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUUGAGGACAAAGAACGAGUGAGAACGAUUGUGAGA (((..(((....)............((..(((((((((((......))))))).))))..))............))...))).......... ( -20.90, z-score = -1.83, R) >droSim1.chr3L 10285870 88 - 22553184 GUGAAACGGCAACAAAAGCAAAUAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUUGAGGACAAAGAACGAGUGAGAAC----GUGAGA ......((((.......))......((..(((((((((((......))))))).))))..)).........))........(----....). ( -19.90, z-score = -1.71, R) >droSec1.super_0 3111016 88 - 21120651 GUGAAACGGCAACAAAAUCAAAUAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUUGAGGACAAAGAACGAGUGAGAAC----GUGAGA ((...(((....)............((..(((((((((((......))))))).))))..))............))....))----...... ( -19.60, z-score = -1.55, R) >droYak2.chr3L 10910596 82 - 24197627 GUGAAACUGCAACAAAAGCAAACAACUUAAGCUGGGAUCUAAAAUCGGAUCUCAAGUUUGGGGACAAAGAACGAGUGUGAGA---------- .......(((.......))).....((((.((((((((((......)))))))...((((....)))).....))).)))).---------- ( -18.80, z-score = -1.71, R) >droEre2.scaffold_4784 10894230 82 - 25762168 GUGAAACGGCAAAAAAAGCUAAAAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUUGGGGACAAAGAACGGGUGUGAGA---------- .......(((.......)))....(((((...((((((((......))))))))..((((....))))...)))))......---------- ( -18.50, z-score = -1.91, R) >consensus GUGAAACGGCAACAAAAGCAAAUAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUUGAGGACAAAGAACGAGUGAGAAC____GUGAGA .........................(((((((((((((((......))))))).)))))))).............................. (-16.46 = -16.06 + -0.40)

| Location | 10,891,842 – 10,891,944 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.25 |
| Shannon entropy | 0.61076 |
| G+C content | 0.43747 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -7.90 |
| Energy contribution | -9.07 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10891842 102 - 24543557 GCUGGGAUAACGGCGAAAAAGGAGAAAGAGGAGCUGUUAAACGGCAACAAAAGCAAAUAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUU-------GAGGACAA ((((...(((((((..................)))))))..))))...............((..(((((((((((......))))))).)))).-------.))..... ( -29.87, z-score = -4.34, R) >droSim1.chr3L 10285891 102 - 22553184 GCUGGGAUAACGGCGAAAAAGGCGAAAGAGGAGCUGUGAAACGGCAACAAAAGCAAAUAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUU-------GAGGACAA ((((.....(((((........(....)....)))))....))))...............((..(((((((((((......))))))).)))).-------.))..... ( -30.20, z-score = -3.96, R) >droSec1.super_0 3111037 102 - 21120651 GCUGGGAUAACGGCGAAAAAGGCGAAAGAGGAGCUGUGAAACGGCAACAAAAUCAAAUAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUU-------GAGGACAA ((((.....(((((........(....)....)))))....))))...............((..(((((((((((......))))))).)))).-------.))..... ( -30.20, z-score = -4.08, R) >droYak2.chr3L 10910611 102 - 24197627 GCUUGGAUAACGGCGAAAAAGGCGAUAGAGGAGCUGUGAAACUGCAACAAAAGCAAACAACUUAAGCUGGGAUCUAAAAUCGGAUCUCAAGUUU-------GGGGACAA ((((...((.((.(......).)).))...))))(((.....(((.......))).....(((((((((((((((......))))))).)))))-------))).))). ( -25.70, z-score = -2.29, R) >droEre2.scaffold_4784 10894245 102 - 25762168 GCUUGGAUAACGGCGAAAAAGGCGAUAGCGGAGCUGUGAAACGGCAAAAAAAGCUAAAAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUU-------GGGGACAA ((((.(.((.((.(......).)).)).).))))(((.....(((.......))).....((..(((((((((((......))))))).)))).-------.)).))). ( -28.50, z-score = -2.88, R) >droAna3.scaffold_13337 15801334 92 - 23293914 -------AAAAAACGUGAGAUAAGGAGGAGAAACUGUGAAACUGU----------GAAAACUUGAGCUGAGACCGAAAAUCGGAUCUCGGUUUUGGGUUUCGAGGACAA -------......(....)..............((.(((((((((----------....))..((((((((((((.....))).)))))))))..))))))).)).... ( -25.00, z-score = -2.36, R) >droMoj3.scaffold_6680 3475897 97 + 24764193 ----GCUCAAAUUUAACAAAUUGCGAAAAGUUUCCG-GCUAGGCUUGCAAUCGAACAACACAGCAACUGCGAUU---AUCCAAACAUCAGCUGAAAG----CAGGACAA ----((................)).....((((.((-(((..((((((..............))))..))(((.---........)))))))).)))----)....... ( -11.83, z-score = 2.04, R) >consensus GCUGGGAUAACGGCGAAAAAGGCGAAAGAGGAGCUGUGAAACGGCAACAAAAGCAAAAAACUUGAGCUGGGAUCUAAAAUCGGAUCUCAAGUUU_______GAGGACAA ................................((((.....))))..................((((((((((((......)))))))).))))............... ( -7.90 = -9.07 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:17 2011