
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,879,570 – 10,879,663 |
| Length | 93 |
| Max. P | 0.777771 |

| Location | 10,879,570 – 10,879,663 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 51.26 |
| Shannon entropy | 0.82784 |
| G+C content | 0.34628 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -4.52 |
| Energy contribution | -4.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.87 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10879570 93 + 24543557 --------GAAAUAAAAUCACUA----CUGACAAAAAUCCAGUAGCCUGCCUCUUAACUUUCUCAAAUUCUUUAG-AAGUUCUCU--UUUAUUUCG-AUACGAAUCUAU--- --------(((((((((...(((----(((.........))))))..........(((((.((.((....)).))-)))))...)--)))))))).-............--- ( -13.10, z-score = -2.13, R) >droSim1.chr3L 10273196 95 + 22553184 --------GAAAUAAAAUCACUA----CUGACAAAAAUCCAGGAGCCUGCCUCUUAACUUCCUCAAAUUCUUUAGGAAGUUCGGU--UUUAUUUCG-AUACGAACCUAUA-- --------(((((((((((....----.............(((((.....)))))((((((((.((....)).)))))))).)))--)))))))).-.............-- ( -21.90, z-score = -3.44, R) >droSec1.super_0 3099228 94 + 21120651 --------GAAAUAAAAUCACUA----CUGACAAAAAUCCAGGAGCCUGCCUCUUAACUUCCUCAAAUUCUUUAGGAAGUUAUGU--UUUAUUUCG-AUACGAAA-UAUA-- --------((((((((((.....----............(((....))).....(((((((((.((....)).))))))))).))--)))))))).-........-....-- ( -19.40, z-score = -3.08, R) >droPer1.super_36 71384 112 + 818889 AAAUUUUAUGACUGACAUCCCGGAAGGCCAACGGGUUCCUGCUAGCCAGUGUCGUGAGCCAACGGAAAAUGUUAAAAGUCAAUGCGACAUAUUUGGCAUACAAAAUUAUGUC ........((((((((((.(((...(((...(((....)))...)))..((.(....).)).)))...)))))...)))))....((((((((((.....))))..)))))) ( -26.90, z-score = -0.60, R) >triCas2.ChLG3 21118588 86 + 32080666 -------AAGAUUGAACUAAUUU-----UAAAAAAGUUGCUAUCCCCGGUCCAAAAAUUUUGUAACUUUGUGUGUGGGCUCUUAGUUAUUGUUCUAAA-------------- -------.(((...((((((.((-----((..((((((((.....................)))))))).....))))...)))))).....)))...-------------- ( -9.70, z-score = 0.76, R) >consensus ________GAAAUAAAAUCACUA____CUGACAAAAAUCCAGUAGCCUGCCUCUUAACUUCCUCAAAUUCUUUAGGAAGUUAUGU__UUUAUUUCG_AUACGAAA_UAU___ ........(((((((........................(((....)))...(((((((((((.((.....)))))))))))))....)))))))................. ( -4.52 = -4.92 + 0.40)

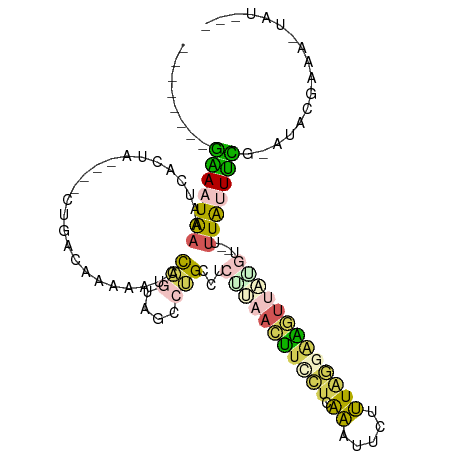
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:15 2011