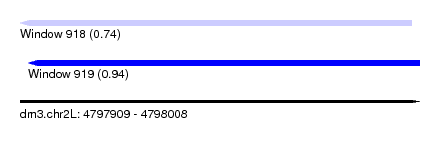
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,797,909 – 4,798,008 |
| Length | 99 |
| Max. P | 0.935205 |

| Location | 4,797,909 – 4,798,006 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 71.64 |
| Shannon entropy | 0.49990 |
| G+C content | 0.51371 |
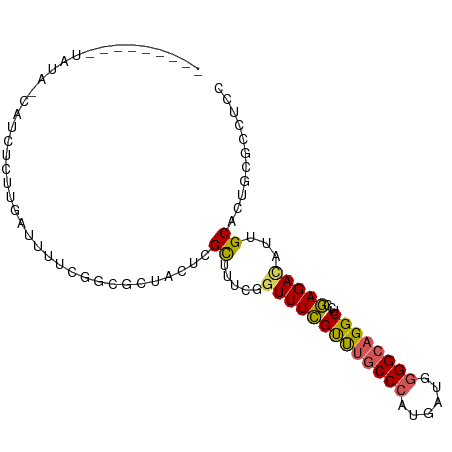
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
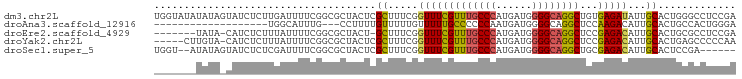
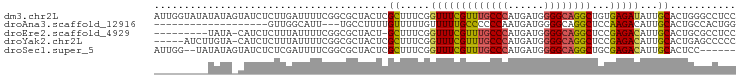
>dm3.chr2L 4797909 97 - 23011544 UGGUAUAUAUAGUAUCUCUUGAUUUUCGGCGCUACUCGCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUGUGAGAUAUUGCACUGGGCCUCCGA .(((.....(((((((((..((...((((.((.....))..))))..))((((((((......))))))))...)))))))))......)))..... ( -29.10, z-score = -0.49, R) >droAna3.scaffold_12916 9366018 75 - 16180835 -------------------UGGCAUUUG---CCUUUUGUUUUUGUUUUUGCCCCCCAAUGAUGGGGCAGGCUCCAAGACAUUGCACUGCCACUGGGA -------------------(((((..((---(....((((((.(..((((((((........))))))))..).))))))..))).)))))...... ( -26.60, z-score = -1.32, R) >droEre2.scaffold_4929 4879100 88 - 26641161 -------UAUA-CAUCUCUUUAUUUUCGGCGCUACU-GCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUCCGAGACAUUGCACUGCGCCUCCGA -------....-...............(((((...(-((((((((....((((((((......)))))))).))))))....)))..)))))..... ( -32.20, z-score = -3.35, R) >droYak2.chr2L 4807289 91 - 22324452 -----CUUGUA-CAUCUCUUUAUUUUCGGCGCUACUCGCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUCCGAGACAUUGCACUGAGCCCCCAA -----......-...............((.(((....((((((((....((((((((......)))))))).))))))....))....))))).... ( -25.90, z-score = -0.98, R) >droSec1.super_5 2893991 89 - 5866729 UGGU--AUAUAGUAUCUCUCGAUUUUCGGCGCUACUCGCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUGCGAGACAUUGCACUCCGA------ .((.--...((((.((((((((.....((((.....)))).))))....((((((((......))))))))...)))).))))....))..------ ( -25.60, z-score = -0.24, R) >consensus _______UAUA_CAUCUCUUGAUUUUCGGCGCUACUCGCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUCCGAGACAUUGCACUGCGCCUCCGA .....................................((.....(((((((((((((......))))))))...)))))...))............. (-15.62 = -15.50 + -0.12)
| Location | 4,797,911 – 4,798,008 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.97 |
| Shannon entropy | 0.51409 |
| G+C content | 0.50951 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
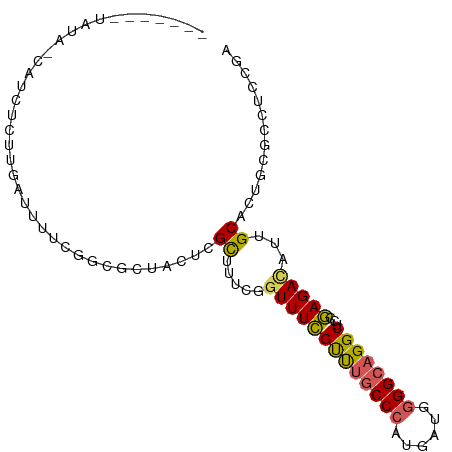
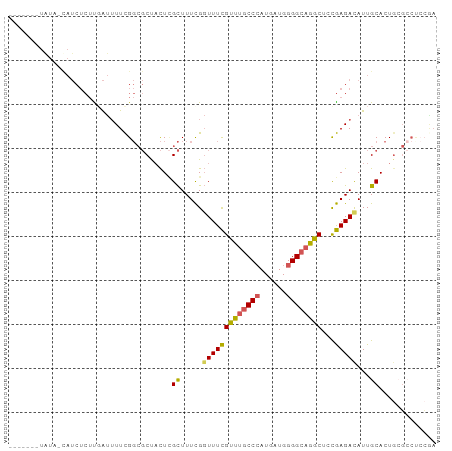
>dm3.chr2L 4797911 97 - 23011544 AUUGGUAUAUAUAGUAUCUCUUGAUUUUCGGCGCUACUCGCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUGUGAGAUAUUGCACUGGGCCUCC ...(((.....(((((((((..((...((((.((.....))..))))..))((((((((......))))))))...)))))))))......)))... ( -29.10, z-score = -0.95, R) >droAna3.scaffold_12916 9366020 75 - 16180835 -------------------GUUGGCAUU---UGCCUUUUGUUUUUGUUUUUGCCCCCCAAUGAUGGGGCAGGCUCCAAGACAUUGCACUGCCACUGG -------------------..(((((..---(((....((((((.(..((((((((........))))))))..).))))))..))).))))).... ( -26.80, z-score = -1.96, R) >droEre2.scaffold_4929 4879102 86 - 26641161 ---------UAUA-CAUCUCUUUAUUUUCGGCGCUACU-GCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUCCGAGACAUUGCACUGCGCCUCC ---------....-...............(((((...(-((((((((....((((((((......)))))))).))))))....)))..)))))... ( -32.20, z-score = -3.83, R) >droYak2.chr2L 4807291 91 - 22324452 -----AUCUUGUA-CAUCUCUUUAUUUUCGGCGCUACUCGCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUCCGAGACAUUGCACUGAGCCCCC -----........-...............((.(((....((((((((....((((((((......)))))))).))))))....))....))))).. ( -25.90, z-score = -1.12, R) >droSec1.super_5 2893993 89 - 5866729 AUUGG--UAUAUAGUAUCUCUCGAUUUUCGGCGCUACUCGCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUGCGAGACAUUGCACUCC------ .....--......(((..(((((......((((.....)))).........((((((((......))))))))..)))))...))).....------ ( -24.30, z-score = -0.35, R) >consensus _________UAUA_CAUCUCUUGAUUUUCGGCGCUACUCGCUUUCGGUUUCGUUUGCCCAUGAUGGGGCAGGCUCCGAGACAUUGCACUGCGCCUCC .......................................((.....(((((((((((((......))))))))...)))))...))........... (-15.62 = -15.50 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:27 2011