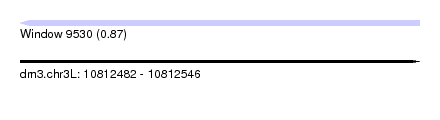
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,812,482 – 10,812,546 |
| Length | 64 |
| Max. P | 0.871825 |

| Location | 10,812,482 – 10,812,546 |
|---|---|
| Length | 64 |
| Sequences | 8 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 63.55 |
| Shannon entropy | 0.74284 |
| G+C content | 0.42972 |
| Mean single sequence MFE | -15.28 |
| Consensus MFE | -5.65 |
| Energy contribution | -5.53 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.82 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
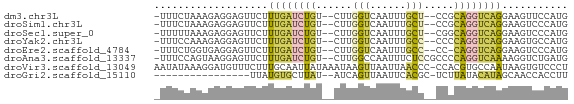
>dm3.chr3L 10812482 64 - 24543557 -UUUCUAAAGAGGAGUUCUUUGAUCUGU--CUUGGUCAAUUUGCU--CCGCAGGUCAGGAAGUUCCAUG -..........(((((((((.(((((((--...(((......)))--..)))))))))))).))))... ( -17.60, z-score = -1.52, R) >droSim1.chr3L 10206958 64 - 22553184 -UUUCUAAAGAGGAGUUCUUUGAUCUGU--CUUGGUCAAUUUGCU--CCGCAGGUCAGGAAGUCCCAUG -........(.(((.(((((.(((((((--...(((......)))--..)))))))))))).))))... ( -19.30, z-score = -2.01, R) >droSec1.super_0 3033140 64 - 21120651 -UUUUUAAAGAGGAGUUCUUUGAUCUGU--CUUGGUCAAUUUGCU--CGGCAGGUCAGGAAGUCCCAUG -........(.(((.(((((.(((((((--(..(((......)))--.))))))))))))).))))... ( -21.50, z-score = -2.91, R) >droYak2.chr3L 10829783 64 - 24197627 -UUUCCAAAGAGGAGUUCUUUGAUCUGU--CUUGGUCAAUUUGCC--CCCCAGGUCAGGAAGUGCCAUG -..........(((.(((((.((((((.--...(((......)))--...))))))))))).).))... ( -15.70, z-score = -0.47, R) >droEre2.scaffold_4784 10816162 63 - 25762168 -UUUCUGGUGAGGAGUUCUUUGAUCUGU--CUUGGUCAAUUUGCC--CC-CAGGUCAGGAAGUCCCAUG -......(((.(((.(((((.((((((.--...(((......)))--..-))))))))))).)))))). ( -20.90, z-score = -1.91, R) >droAna3.scaffold_13337 15724460 66 - 23293914 -UUUCCAGUAAGGAGUUCUUUGAUCUGU--CUUGGCCAAUUUCUCCGCCCCAGGUCAAAAGGUCUGAUG -..........((((....(((..(...--...)..)))...))))....(((..(....)..)))... ( -14.50, z-score = -0.37, R) >droVir3.scaffold_13049 15292301 68 + 25233164 AAUAUAAAGGAUGUUUCUUUGCAAUUAUAAAUAAGUUAAUUAACCC-CCACGUGCCAAUAAGUGUCCCU ........(((..(((.((.(((........(((.....)))....-.....))).)).)))..))).. ( -7.73, z-score = -0.82, R) >droGri2.scaffold_15110 4052094 50 + 24565398 ----------------UUAUGUGCUUAU--AUCAGUUAAUUCACGC-UCUUAUACAUAGCAACCACCUU ----------------.....(((((((--(..(((........))-)..))))...))))........ ( -5.00, z-score = -0.98, R) >consensus _UUUCUAAAGAGGAGUUCUUUGAUCUGU__CUUGGUCAAUUUGCC__CCGCAGGUCAGGAAGUCCCAUG ...................((((((((......(((......))).....))))))))........... ( -5.65 = -5.53 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:05 2011