| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,797,323 – 10,797,428 |
| Length | 105 |
| Max. P | 0.989507 |

| Location | 10,797,323 – 10,797,428 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 65.09 |
| Shannon entropy | 0.64159 |
| G+C content | 0.36936 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -11.05 |
| Energy contribution | -11.01 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
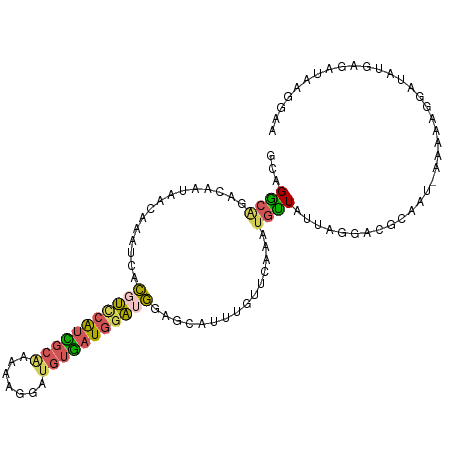
>dm3.chr3L 10797323 105 - 24543557 GCAGGCAGACAAUAACAAAUCACGUUCAUCGCAAAAAGGAUGUGAUGGAUGGAGCAUUUGUUCAAAUGCUAUUAGGACUCAAU-GAAAAGGAUAUGAAAUAAGGAA ...............((.(((.(((((((((((.......))))))))))).(((((((....))))))).............-......))).)).......... ( -22.70, z-score = -1.95, R) >droAna3.scaffold_13337 15708598 106 - 23293914 CCAGACAACCAAUAACAAAUAAUGCAAGAGCUUACAAUACCCUUAUAAGAAGGGAUUAAUUCGAAACUUUGUUUCAGAAGAGUCUAUAAAAAUGGGAAAUAAAAAA ..((((.............((((((....))........(((((.....))))))))).((((((((...))))).)))..))))..................... ( -14.30, z-score = -0.64, R) >droEre2.scaffold_4784 10800197 105 - 25762168 GCAGGCAGACAAUAACAAAUCACGUCCAUUGCGAAAAGGAUGUGAUGGAUGGAACAUUUUCUCAAAUGUUAUUAGGACGCCAU-AAGAAGGAUAUGAGAGAAGGAA ...(((...((((((((.(((((((((.((....)).)))))))))...(((((....))).))..))))))).)...)))..-...................... ( -23.00, z-score = -1.74, R) >droYak2.chr3L 10813335 87 - 24197627 GCAGGCAGACAAUAACAAAUCACGUCCAUCGCAAAAAGGAUGUGAUGGAAGGAA--------CAAAUGUUAUUAUGAAGUA---UAUUAGGUUAAGAG-------- ((...((...(((((((..((...(((((((((.......)))))))))..)).--------....))))))).))..)).---..............-------- ( -17.30, z-score = -2.31, R) >droSec1.super_0 3017287 106 - 21120651 GCAGGCGGACCAUAACAAAUCACGUUCAUCGCAAAAAGGAUGUGAUGGAUGGAGCAUUUGUUCAAAUGCUAUUAGGACUCCUUCAAAAAGGAUAUGAGAUAAGGAA .........((...........(((((((((((.......))))))))))).(((((((....)))))))....))..(((((....))))).............. ( -25.80, z-score = -1.52, R) >consensus GCAGGCAGACAAUAACAAAUCACGUCCAUCGCAAAAAGGAUGUGAUGGAUGGAGCAUUUGUUCAAAUGUUAUUAGGACGCAAU_AAAAAGGAUAUGAGAUAAGGAA ...((((...............(((((((((((.......)))))))))))...............)))).................................... (-11.05 = -11.01 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:03 2011