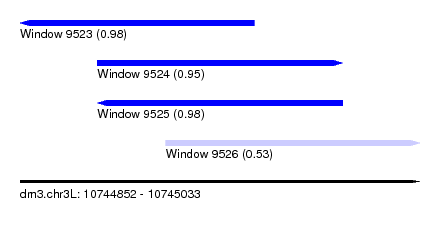
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,744,852 – 10,745,033 |
| Length | 181 |
| Max. P | 0.980666 |

| Location | 10,744,852 – 10,744,958 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.88 |
| Shannon entropy | 0.57650 |
| G+C content | 0.34163 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -8.57 |
| Energy contribution | -9.92 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10744852 106 - 24543557 -----AUUUCCGAACUCAAGCUAUU--AAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUCAGCCUUCCUCAGGAGCUUUCAUUUGCCCAUUGGCAA -----......((..((((((((..--.(((((((....)))))))....))))))))..))................(((.......((.((........))))...))).. ( -24.10, z-score = -3.41, R) >droSim1.chr3L 10138191 106 - 22553184 -----AUUGCCGAACUCAAGCUAUU--AAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUUGCCUGCCUCAGGAGCUUUCAUUUGGCCAUUGGCAA -----.(((((((..((((((((..--.(((((((....)))))))....)))))))).........(((((.......((((...)))).......)))))....))))))) ( -30.54, z-score = -3.99, R) >droSec1.super_0 2965059 106 - 21120651 -----AUUGCCGAACUCAAGCUAUU--AAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUUGCCUGCCUCAGGAGCUUUCAUUUGGCCAUUGGCAA -----.(((((((..((((((((..--.(((((((....)))))))....)))))))).........(((((.......((((...)))).......)))))....))))))) ( -30.54, z-score = -3.99, R) >droYak2.chr3L 10757261 92 - 24197627 -----------UAUUUCAAGUUAUU--AAAUUCCUUCUACGGUAUACACUUAGUUUGAAAUUCUUUACAAAUUAUGUAGCAUUCGCCAGGAGCUUUAA--UGGCCAA------ -----------........((((((--((((((((....(((..((((..(((((((.((....)).)))))))))))....)))..))))).)))))--))))...------ ( -18.00, z-score = -2.06, R) >droWil1.scaffold_180949 109932 111 + 6375548 GUGGAACAGAUGUUGUUAACUUUUUUGAAAAUUGUUCCAUUAUUAAAAUUUA-CUUGAGUUUUUGCACGUAUAGUUUUGUGUGUUGCUUCAAUUUGAAGUUUGAAAUUUUCA- (((((((((.(....((((.....)))).).)))))))))............-..((((..((((((((........)))))...(((((.....)))))...)))..))))- ( -18.20, z-score = -0.01, R) >consensus _____AUUGCCGAACUCAAGCUAUU__AAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUUGCCUGCCUCAGGAGCUUUCAUUUGGCCAUUGGCAA ........(((((..((((((((.....(((((((....)))))))....))))))))....................((((......)).)).............))))).. ( -8.57 = -9.92 + 1.35)

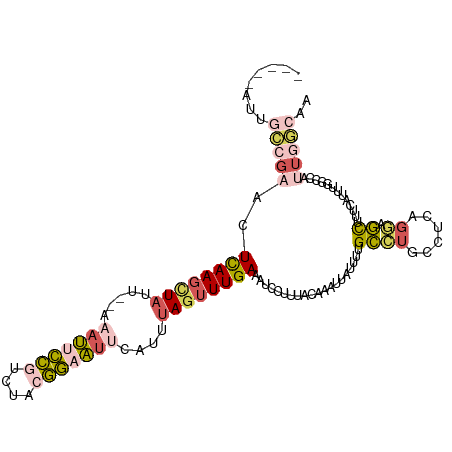
| Location | 10,744,887 – 10,744,998 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.43 |
| Shannon entropy | 0.28964 |
| G+C content | 0.29223 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -14.59 |
| Energy contribution | -16.28 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10744887 111 + 24543557 UGAAUAAUUUGUAAAGGAUUUCAAACUAAAUGAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGAAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGCA .........(((((((..((((........(((((((((....))))))))).........(((((.((((((.....)))))).)))))......))))..))..))))) ( -30.20, z-score = -4.46, R) >droSim1.chr3L 10138226 111 + 22553184 AAAAUAAUUUGUAAAGGAUUUCAAACUAAAUGAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGAA .............(((..((((........(((((((((....))))))))).........(((((.((((((.....))).))))))))......))))..)))...... ( -25.20, z-score = -2.85, R) >droSec1.super_0 2965094 111 + 21120651 AAAAUAAUUUGUAAAGGAUUUCAAACUAAAUGAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGCAAGAAAGACUUCUUGAA ....................((((.(((..(((((((((....))))))))).))).))))(((((.((((((.....))).))))))))...((((((....)))))).. ( -27.40, z-score = -2.94, R) >droYak2.chr3L 10757288 88 + 24197627 UACAUAAUUUGUAAAGAAUUUCAAACUAAGUGUAUACCGUAGAAGGAAUUUAAUAACUUGA------------AAUAAUCAUUUGCGAGCAUAGAAUUAA----------- .......(((((((((((((((((((.....))...((......))...........))))------------)))..)).))))))))...........----------- ( -10.70, z-score = -0.31, R) >consensus AAAAUAAUUUGUAAAGGAUUUCAAACUAAAUGAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGAA .......((((((((.((((((((.(((..(((((((((....))))))))).))).)))).((((....))))..)))).))))))))...................... (-14.59 = -16.28 + 1.69)

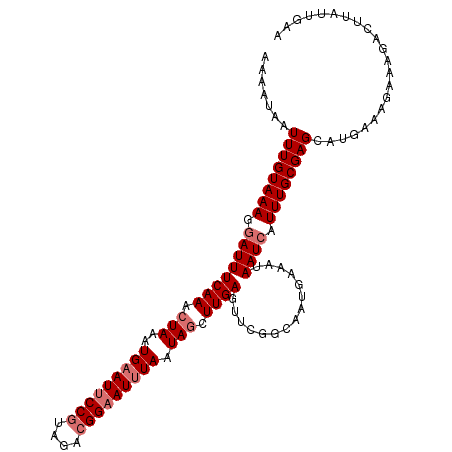
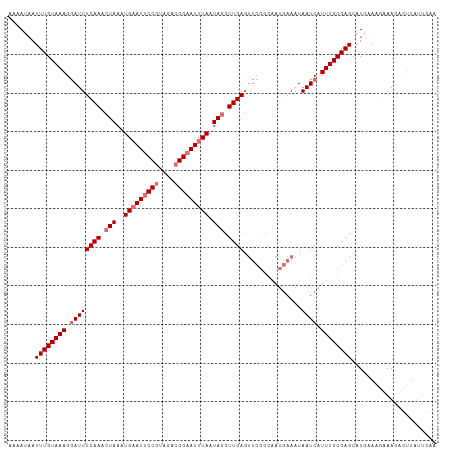
| Location | 10,744,887 – 10,744,998 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.43 |
| Shannon entropy | 0.28964 |
| G+C content | 0.29223 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -12.11 |
| Energy contribution | -12.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10744887 111 - 24543557 UGCAAUAAGUCUUUCUUUCAUGCUCGCAAAUGAUUAUUUCAUUUCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUCA .(((..(((.....)))...)))(((.((((((.....)))))).)))..((((((((...(((((((....)))))))....)))))))).................... ( -25.80, z-score = -5.35, R) >droSim1.chr3L 10138226 111 - 22553184 UUCAAUAAGUCUUUCUUUCAUGCUCGCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUU ...(((((...............(((..(((((.....)))))..)))..((((((((...(((((((....)))))))....)))))))).............))))).. ( -21.70, z-score = -3.64, R) >droSec1.super_0 2965094 111 - 21120651 UUCAAGAAGUCUUUCUUGCAUGCUCGCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUU ..((((((....)))))).....(((..(((((.....)))))..)))..((((((((...(((((((....)))))))....)))))))).................... ( -24.70, z-score = -3.60, R) >droYak2.chr3L 10757288 88 - 24197627 -----------UUAAUUCUAUGCUCGCAAAUGAUUAUU------------UCAAGUUAUUAAAUUCCUUCUACGGUAUACACUUAGUUUGAAAUUCUUUACAAAUUAUGUA -----------.......(((((.((..(((((((...------------...)))))))............))))))).((.(((((((.((....)).))))))).)). ( -6.84, z-score = 0.61, R) >consensus UUCAAUAAGUCUUUCUUUCAUGCUCGCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUA ..................................................((((((((...(((((((....)))))))....)))))))).................... (-12.11 = -12.68 + 0.56)

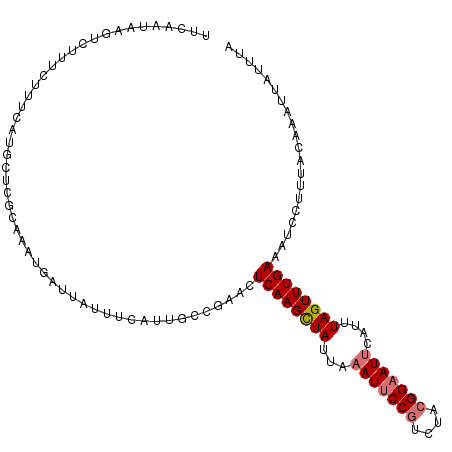
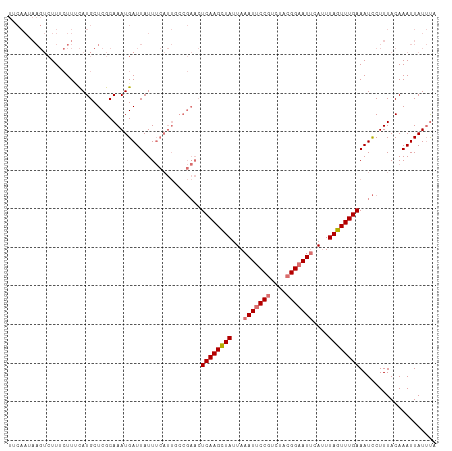
| Location | 10,744,918 – 10,745,033 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.36 |
| Shannon entropy | 0.57217 |
| G+C content | 0.35949 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -7.30 |
| Energy contribution | -10.05 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10744918 115 + 24543557 GAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGAAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGCAAUUGUAAUGAGCACGGCAGCAAGGACCUGAAACCU ((((((((....))))))))....(((...(((((.((((((.....)))))).)))))..........(.((((((((....)))))))))..)))....(((........))) ( -32.70, z-score = -3.77, R) >droSim1.chr3L 10138257 115 + 22553184 GAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGAAACUGGAAUGAGUAGGGAUGCAAGUACCCAAAACCU ((((((((....))))))))....(((((.(((((.((((((.....))).))))))))...........(((((((........))))))).......)))))........... ( -26.80, z-score = -2.15, R) >droSec1.super_0 2965125 115 + 21120651 GAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGCAAGAAAGACUUCUUGAAAUUGGAAUGAGUAGGGAUGCAAGUACCCAAAACCU ((((((((....))))))))....((((..(((((.((((((.....))).))))))))...((((((....))))))..........)))).(((.((....)))))....... ( -28.30, z-score = -1.89, R) >droWil1.scaffold_180949 110006 88 - 6375548 GAACAAUUUUCAAAAAAGUUAACAACAUCUGUUCCAC-----AUUAGCUAUUAGC-------AAAAAAUAUCUUUACGUAAAUAAGAUGU-CAAACCGAUA-------------- ..........................(((.(((....-----....((.....))-------.....(((((((.........)))))))-..))).))).-------------- ( -6.60, z-score = -0.00, R) >consensus GAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGAAAUUGGAAUGAGCAGGGAUGCAAGUACCCAAAACCU ((((((((....))))))))..........(((((..(((((.....)))))..)))))........................................................ ( -7.30 = -10.05 + 2.75)

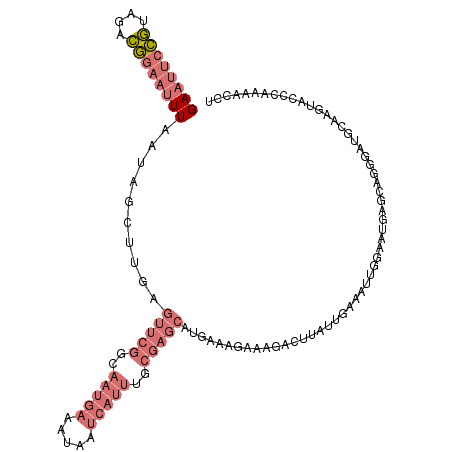
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:17:02 2011