| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,743,637 – 10,743,737 |
| Length | 100 |
| Max. P | 0.539219 |

| Location | 10,743,637 – 10,743,737 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.78 |
| Shannon entropy | 0.62089 |
| G+C content | 0.46954 |
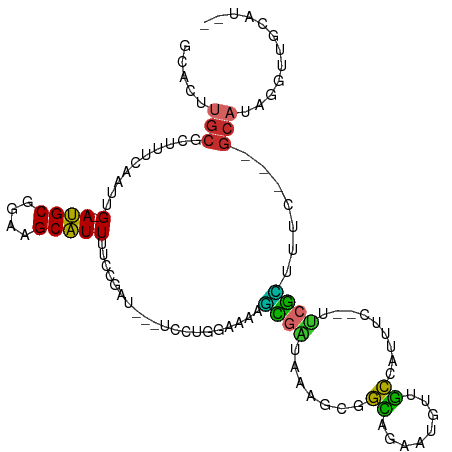
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -10.22 |
| Energy contribution | -8.85 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.85 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.539219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
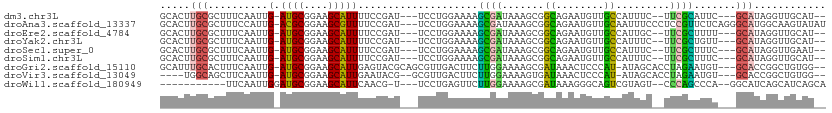
>dm3.chr3L 10743637 100 + 24543557 GCACUUGCGCUUUCAAUUG-AUGCGGAAGCAUUUUCCGAU---UCCUGGAAAAGCGAUAAAGCGGCAGAAUGUUGCCAUUUC--UUCGCAUUC---GCAUAGGUUGCAU-- (((((((.((.........-.(((....)))(((((((..---...)))))))((((..(((.(((((....))))).))).--.))))....---)).)))).)))..-- ( -32.30, z-score = -1.56, R) >droAna3.scaffold_13337 15656646 107 + 23293914 GCACUUGCGCUUUCCAUUG-ACGCGGAAGCGUUUUCCGAU---UCCUGGAAAAGCGAUAAAGCGGCAGAAUGUUGCAAUUUCCCUCCGUUCUCAGGGCAUGGCAAGUAUAU ..((((((.(...((.(((-(.(((((.((.(((((((..---...)))))))))......(((((.....)))))........)))))..))))))...))))))).... ( -33.30, z-score = -0.98, R) >droEre2.scaffold_4784 10746223 100 + 25762168 GCACUUGCGCUUUCAAUUG-AUGCGGAAGCAUUUUCCGAU---UCCUGGAAAAGCGAUAAAGCGGCAGAAUGUUGCCAUUGC--UUCGCUUUU---GCAUAGGUUGCAU-- ((....)).....((((..-((((((((((.(((((((..---...)))))))(((((...(((((.....))))).)))))--...))))))---))))..))))...-- ( -33.60, z-score = -1.61, R) >droYak2.chr3L 10755922 100 + 24197627 GCACUUGCGCUUUCAAUUG-AUGCGGAAGCAUUUUCCGAU---UCCUGGAAAAGCGAUAAAGCGGCAGAAUGUUGCCAUUUC--UUCGCUGUU---GCAUAGGUUGCAU-- (((((((.((........(-((((....)))))(((((..---...))))).(((((..(((.(((((....))))).))).--.)))))...---)).)))).)))..-- ( -32.60, z-score = -1.46, R) >droSec1.super_0 2963909 100 + 21120651 GCACUUGCGCUUUCAAUUG-AUGCGGAAGCAUUUUCCGAU---UCCUGGAAAAGCGAUAAAGCGGCAGAAUGUUGCCAUUUC--UUCGCUUUC---GCAUAGGUUGAAU-- ((....))...((((((..-((((((((((.(((((((..---...)))))))((......))(((((....))))).....--...))))))---))))..)))))).-- ( -37.30, z-score = -3.33, R) >droSim1.chr3L 10137031 100 + 22553184 GCACUUGCGCUUUCAAUUG-AUGCGGAAGCAUUUUCCGAU---UCCUGGAAAAGCGAUAAAGCGGCAGAAUGUUGCCAUUUC--UUCGCUUUC---GCAUAGGUUGCAU-- ((....)).....((((..-((((((((((.(((((((..---...)))))))((......))(((((....))))).....--...))))))---))))..))))...-- ( -34.20, z-score = -2.15, R) >droGri2.scaffold_15110 3975057 104 - 24565398 GCAUUUGCACUUUCAAUUG-AUGCGGAAGCAUUGAGUACGCAGCGUUGACUUCUUGGAAAAGCGAUAAACUCCCAU-AUAGCACCUAGAAUGU---GCACCGGCUGUGG-- ((....))........(..-((((....))))..)...(((((((((.....(((....))).....)))......-...((((.......))---))....)))))).-- ( -28.70, z-score = -0.92, R) >droVir3.scaffold_13049 15216737 98 - 25233164 ----UGGCAGCUUCAAUUG-AUGCGGAAGCAUUGAAUACG--GCGUUGACUUCUUGGAAAAGUGAUAAACUCCCAU-AUAGCACCUAGAAUGU---GCACCGGCUGUGG-- ----..(((((.....(..-((((....))))..)...((--(.(((.....(((....))).....)))......-...((((.......))---)).))))))))..-- ( -28.80, z-score = -1.59, R) >droWil1.scaffold_180949 108654 92 - 6375548 -----------UUCAAUUGGAUGCGGAAGCAUUCAACG-U---UCCUGAGUUCUUGGAAAAGCGAUAAAGGGCAGUCGUAGU--CCCAGCCCA--GGCAUCAGCAUCAGCA -----------.....((((((((....))))))))..-.---.((((.(((...(((...(((((........)))))..)--)).))).))--)).....((....)). ( -27.60, z-score = -1.65, R) >consensus GCACUUGCGCUUUCAAUUG_AUGCGGAAGCAUUUUCCGAU___UCCUGGAAAAGCGAUAAAGCGGCAGAAUGUUGCCAUUUC__UUCGCUUUC___GCAUAGGUUGCAU__ .....(((............((((....)))).....................((((.......((........)).........)))).......)))............ (-10.22 = -8.85 + -1.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:56 2011