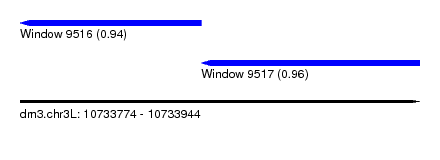
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,733,774 – 10,733,944 |
| Length | 170 |
| Max. P | 0.957347 |

| Location | 10,733,774 – 10,733,851 |
|---|---|
| Length | 77 |
| Sequences | 7 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Shannon entropy | 0.35291 |
| G+C content | 0.54216 |
| Mean single sequence MFE | -23.64 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938616 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
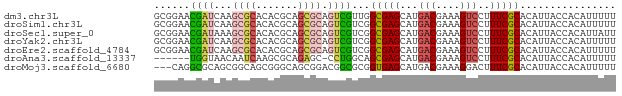
>dm3.chr3L 10733774 77 - 24543557 GCGGAACGAUCAAGCGCACACGCAGCGCAGUCGUUGGCGAGCAUGACGAAAGUCCUUUCGCACAUUACCACAUUUUU (.((((((((...((((.......)))).)))))).(((((...(((....)))..)))))......)).)...... ( -25.40, z-score = -2.64, R) >droSim1.chr3L 10127065 77 - 22553184 GCGGAACGAUCAAGCGCACACGCAGCGCAGUCGUCGGCGAGCAUGACGAAAGUCCUUUCGCACAUUACCACAUUUUU (.((.(((((...((((.......)))).)))))..(((((...(((....)))..)))))......)).)...... ( -23.80, z-score = -2.01, R) >droSec1.super_0 2954328 77 - 21120651 GCGGAACGAUAAAGCGCACACGCAGCGCAGUCGUCGGCGAGCAUGACGAAAGUCCUUUCGCACAUUACCACAUUAUU (.((.(((((...((((.......)))).)))))..(((((...(((....)))..)))))......)).)...... ( -23.80, z-score = -2.14, R) >droYak2.chr3L 10745702 77 - 24197627 GCGGAACGAUCAAGCGCACACGCAGCGCAGUCGUCGGCGAGCAUGACGAAAGUCCUUUCGCACAUUACCACAUUUUU (.((.(((((...((((.......)))).)))))..(((((...(((....)))..)))))......)).)...... ( -23.80, z-score = -2.01, R) >droEre2.scaffold_4784 10736127 77 - 25762168 GCGGAACGAUCAAGCGCACACGCAGCGCAGUCGUCGGCGAGCAUGACGAAAGUCCUUUCGCACAUUACCACAUUUUU (.((.(((((...((((.......)))).)))))..(((((...(((....)))..)))))......)).)...... ( -23.80, z-score = -2.01, R) >droAna3.scaffold_13337 15646966 70 - 23293914 ------UGGUAACAAUCAAGCGCAGAGC-CCUGGCAGCGAGCAUGACGAAAGUCCUUUCGCACAUUACCACAUUUUU ------((((((.......((.(((...-.))))).(((((...(((....)))..)))))...))))))....... ( -22.30, z-score = -3.59, R) >droMoj3.scaffold_6680 10758812 74 - 24764193 ---CAGGCGCAGCGGCAGCGGGCAGCGGACGGCGCGGUGAGCAUGACGAAAGGACUUUCGCACAUUACCACAUUUUU ---...((((..(.((........)).)...))))(((((...((.(((((....)))))))..)))))........ ( -22.60, z-score = -1.01, R) >consensus GCGGAACGAUCAAGCGCACACGCAGCGCAGUCGUCGGCGAGCAUGACGAAAGUCCUUUCGCACAUUACCACAUUUUU ......((((...((((.......)))).))))...(((((...(((....)))..)))))................ (-15.15 = -15.50 + 0.35)
| Location | 10,733,851 – 10,733,944 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 67.84 |
| Shannon entropy | 0.52679 |
| G+C content | 0.32522 |
| Mean single sequence MFE | -14.48 |
| Consensus MFE | -8.58 |
| Energy contribution | -8.42 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
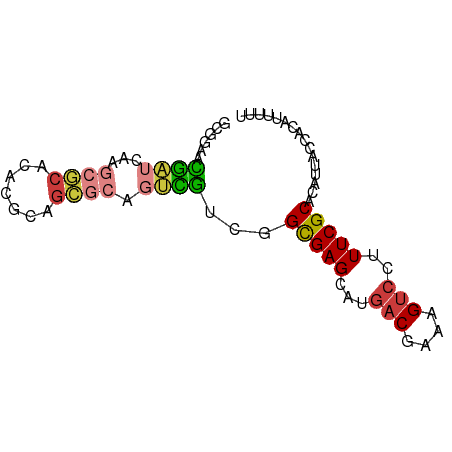
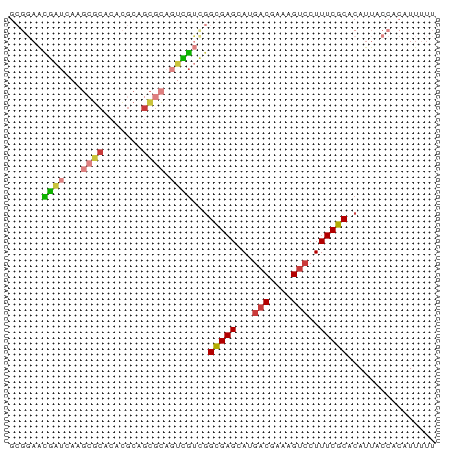
>dm3.chr3L 10733851 93 - 24543557 -----------AUCUAAUUCAAAUCUAAGUGAUUAAAGCUUAGCUUUGUAGUCACAA-UUUU--UUUCUUAACUUCGUAAUACCCUUCUUUUCGCCAUAGGGUAUUU -----------.................(((((((((((...))))..)))))))..-....--..............((((((((............)))))))). ( -16.90, z-score = -2.94, R) >droSim1.chr3L 10127142 84 - 22553184 -------------------CAAAUCUAAGUUAUUCAAGCUUAGCUUUGCAGUCAAUA--UUC--UUCCUUAAGUUCGUGAUACCCUUCUUUUUGCCAUAGGGUAUUU -------------------((((.(((((((.....))))))).)))).........--...--..............((((((((............)))))))). ( -16.10, z-score = -2.28, R) >droSec1.super_0 2954405 86 - 21120651 -------------------CAAAUCUAAGUUAUUUAAGCUUAGCUUUGCAGUCACAAUAUUC--UUCCUUAAGUUCGUGAUACCCUUCUUUUCGCCAUAGGGUAUUU -------------------((((.(((((((.....))))))).))))..............--..............((((((((............)))))))). ( -16.10, z-score = -2.33, R) >droYak2.chr3L 10745779 84 - 24197627 -----------------------CUUAAAUUUUUAAGCAUUUUCAUUAAAGCUGUCAUAGUUAGUUUUCUAAGUUCGUGAUACCCUUUUUCUUGCCAUAGGGUAUUU -----------------------(((((....)))))..........(((((((.......)))))))..........((((((((............)))))))). ( -11.80, z-score = -0.80, R) >droEre2.scaffold_4784 10736204 105 - 25762168 CGUAGAUAUUAAAACAUUUCCCAGCAAAGUCUUUAAAUGUUAUUCUUGCAGCCAUAAUAUUU--UUCCCCAAGUUCGUGAUACCCUUUUUUUUGCCAUAGGGUAUUU ...((((((((.(((((((...((......))..))))))).....((....))))))))))--..............((((((((............)))))))). ( -11.50, z-score = 0.36, R) >consensus ___________________CAAAUCUAAGUUAUUAAAGCUUAGCUUUGCAGUCACAAUAUUU__UUCCUUAAGUUCGUGAUACCCUUCUUUUUGCCAUAGGGUAUUU ..............................................................................((((((((............)))))))). ( -8.58 = -8.42 + -0.16)

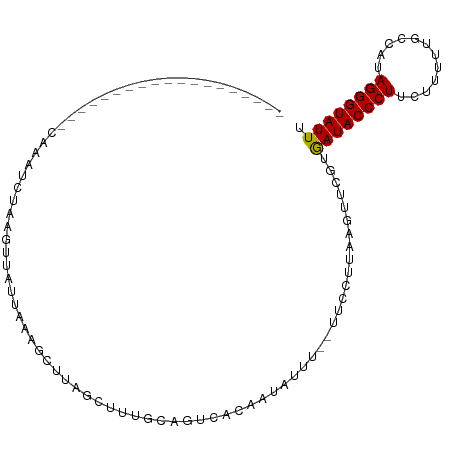
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:55 2011