| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,787,534 – 4,787,627 |
| Length | 93 |
| Max. P | 0.642901 |

| Location | 4,787,534 – 4,787,627 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 64.14 |
| Shannon entropy | 0.63510 |
| G+C content | 0.38736 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -8.92 |
| Energy contribution | -8.41 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
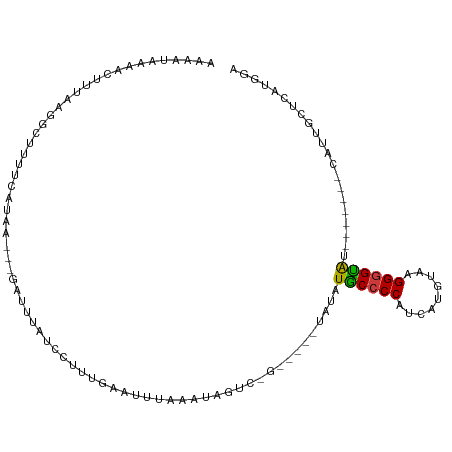
>dm3.chr2L 4787534 93 - 23011544 AAAAUAUAACUUUAAGGCAUUUCAUAG---GAUAUUUCCUUUGAAUUUAAAUGGUC-G-----UAUAUACCCCAUCAUGUAAGGAGUAA-------CAUUGCUCAUGGA ...(((..(((.........((((.((---((....)))).)))).......))).-.-----))).....((((........(((((.-------...))))))))). ( -15.89, z-score = -0.11, R) >droPer1.super_8 1705326 100 + 3966273 -AUGUGAAAACCCAAUACUUUUCGAGAAUCGAGCUAUUCUGGGAACUUACUUAGUUCUU-ACAUAUAUGCCCCACCACCUAAGGGGCGU-------CAUGCCCUAAGAG -..((((...((((......((((.....))))......))))...)))).....((((-(..((((((((((.........)))))))-------.)))...))))). ( -23.80, z-score = -1.55, R) >dp4.chr4_group3 658629 100 + 11692001 -AUGUGACAACCCAAUACUUUUCGAGAAUCGAGCUAUUCUGGGAACUUCCUUAGUUCUU-ACAUAUAUGCCCCACCACCUAAGGGGCGU-------CAUGCCCUAAGAG -..(((((...(((......((((.....))))......)))(((((.....)))))..-........(((((.........)))))))-------))).......... ( -24.40, z-score = -1.63, R) >droEre2.scaffold_4929 4863328 106 - 26641161 AGUAUAAAACUUUGCGACAUUUUAUGC---GAUUUAUCUUUUGCACUUCAAUCGUCUGUAACCUAUAUGCCCCAUCCUGUAAGGGGUAAGGGGUACCAUUGCUCAUGGA ...........((((((((((...(((---((........)))))....))).))).))))......(((((((((((....)))))..))))))((((.....)))). ( -24.20, z-score = -0.37, R) >droYak2.chr2L 4796548 91 - 22324452 AAAAUAAAACUUCAAGGCACU---UAA---GACUUAUGAUUUGAAUUUACAUCGUCAG-----CAUAUACCCCAACCAGUAAGGGGUAG-------CAUUGCUCAUGGA ..........((((((.((..---...---......)).))))))......((((.((-----((..((((((.........)))))).-------...)))).)))). ( -18.30, z-score = -1.05, R) >droSec1.super_5 2883824 93 - 5866729 AAAAUAAAACCAUGAGGCUUUCUAUAG---GAUUUUUCCUUUGAAUUUAAAUUGUC-G-----UAUAUACCCCAUCGUGUAAGGGGUAU-------CAGUGCUCAUGGA .........(((((((((((((...((---((....))))..)))...........-.-----...(((((((.........)))))))-------.))).))))))). ( -27.00, z-score = -3.12, R) >droSim1.chr2L 4656940 93 - 22036055 AAAAUAAAACUGUAAGGCUUUCUAUAG---GAUUUUUCCUUUGAAUUUAAAUGGUC-G-----UAUAUGCCCCAUCGUGUAAGGGGUAU-------CAGUGCUCAUGGA .........((((..(((((((...((---((....))))..))).......))))-(-----((((((((((.........)))))))-------..))))..)))). ( -20.31, z-score = -0.71, R) >consensus AAAAUAAAACUUUAAGGCUUUUCAUAA___GAUUUAUCCUUUGAAUUUAAAUAGUC_G_____UAUAUGCCCCAUCAUGUAAGGGGUAU_______CAUUGCUCAUGGA ...................................................................((((((.........))))))..................... ( -8.92 = -8.41 + -0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:25 2011