| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,698,906 – 10,699,007 |
| Length | 101 |
| Max. P | 0.985454 |

| Location | 10,698,906 – 10,699,007 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.78 |
| Shannon entropy | 0.66996 |
| G+C content | 0.50836 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -9.81 |
| Energy contribution | -11.33 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10698906 101 + 24543557 ------ACACACAUACACGCUGGGUCCAGGAUCCC--UUUUAACCCGGGAUUCUGCACCACCAUCUUCC-----UUCCAUCCUCUCCUCUCCACCAGUCGAGUAUUAAGUAAAU ------.......(((......(((.(((((((((--.........))))))))).)))..........-----.........(((..((.....))..)))......)))... ( -21.50, z-score = -2.11, R) >droSim1.chr3L 10091305 101 + 22553184 ------ACACACAUACACGCAGGGUCCAGGAUCCC--UUUUAACCCGGGAUUCUGCACCACCAUCGGCC-----UCCGCUCCUCUCGUCUCCGCCAGUCGAGUAUUAAGUAAAU ------.......(((..((..(((.(((((((((--.........))))))))).)))......((..-----.))))....((((.((.....)).))))......)))... ( -26.40, z-score = -1.98, R) >droSec1.super_0 2919709 101 + 21120651 ------GCACACAAACACGCAGGGUCCAGGAUCCC--UUUUAACCCGGGAUUCUGCACCACCAUCGGCC-----UCCACUCCUCUCGUCUCCGCCAGUCGAGUAUUAAGUAAAU ------...........((...(((.(((((((((--.........))))))))).))).....))...-----...(((...((((.((.....)).)))).....))).... ( -24.20, z-score = -1.51, R) >droYak2.chr3L 10709242 94 + 24197627 ------ACACACAUACACGCAGGGUCCAGGAUCCC--UUUUAACCCGGGAUUCUGCACCACCUC------------CACUUCGCUCGACUCCUCCACUCGAGUAUUGAGUAAAU ------................(((.(((((((((--.........))))))))).))).....------------.((((.((((((.........))))))...)))).... ( -25.80, z-score = -3.03, R) >droEre2.scaffold_4784 10697845 106 + 25762168 ------ACACACAUACACGCAGGGUCCAGGAUCCC--UUUUAACCCGGGAUUCUGCACCACCUCUUGCUCUGGGCUCCUUCCCCUCGACUCCUCCAGUCGAGUAUUGAGUAAAU ------................(((.(((((((((--.........))))))))).))).....((((((.(((......)))(((((((.....)))))))....)))))).. ( -35.60, z-score = -3.51, R) >droAna3.scaffold_13337 15610632 80 + 23293914 ------ACACACACACACGCAAA---AAGGAUC-----UGCAAAUGGGGGACCAGGAGGA-----GUCC-----UUUUGGUUCCA----------CCUCGAACAUUAAGUAAAU ------............(((..---.......-----)))....((((((((((((((.-----..))-----)))))))))).----------))................. ( -21.40, z-score = -1.83, R) >dp4.chrXR_group6 6979788 106 - 13314419 ACACACACACACACAGACACAGA---CAGAAUCGCAUCUACAGGAUGCGGAGUACGGGCACAAAUAUCC-----UCUUCCAUCUGUAACCUCAUGCUUCGUGCAUUAAGUAAAU ...............((.....(---((((...(((((.....)))))((((...(((........)))-----..)))).)))))....))((((.....))))......... ( -20.60, z-score = -1.01, R) >consensus ______ACACACAUACACGCAGGGUCCAGGAUCCC__UUUUAACCCGGGAUUCUGCACCACCAUCGGCC_____UCCACUCCCCUCGACUCCACCAGUCGAGUAUUAAGUAAAU ......................(((.(((((((((...........))))))))).)))....................................................... ( -9.81 = -11.33 + 1.51)

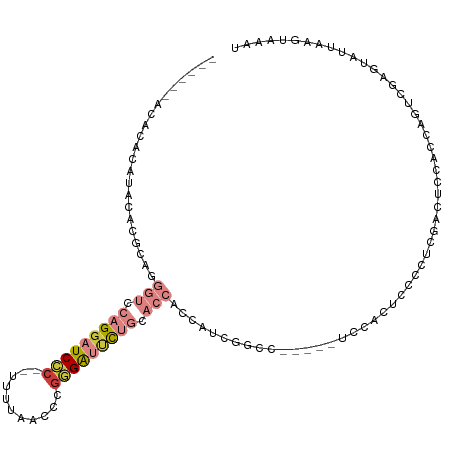
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:51 2011