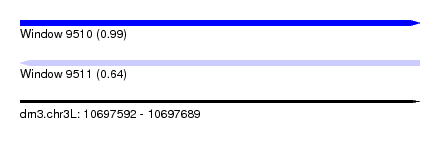
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,697,592 – 10,697,689 |
| Length | 97 |
| Max. P | 0.986611 |

| Location | 10,697,592 – 10,697,689 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.12 |
| Shannon entropy | 0.28982 |
| G+C content | 0.35887 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.36 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
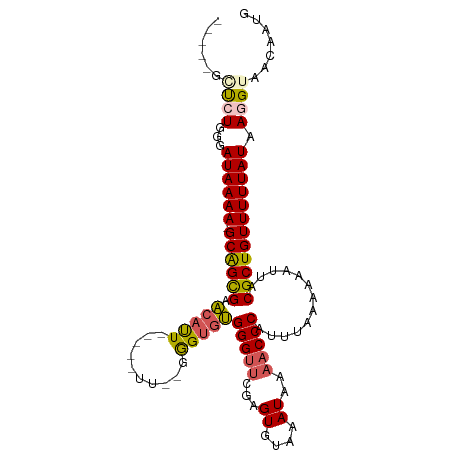
>dm3.chr3L 10697592 97 + 24543557 ---CUGGCGUGGUAUAAAAAGCAGCGUACAUU-----UU--GGGUGUGGGUUCGAGUGUAAAUAAAACCCAUUUAAAAAAUUACGCUGUUUUUAUAAGGUAACAAUG ---.....((..((((((((.(((((((.(((-----((--....(((((((..(.......)..)))))))....)))))))))))))))))))).....)).... ( -26.60, z-score = -2.82, R) >droSec1.super_0 2918363 96 + 21120651 ---CUGGCCUCGUAUAAAA-GCAGCGAACAUU-----UU--UGGUGUGGGUUCGAGUGUAAAUAAAACCCAUUUAAAAAAUUACGCUGUUUUUAUAAGGUAACAAUG ---...((((..(((((((-((((((...(((-----((--(...(((((((..(.......)..)))))))...))))))..)))))))))))))))))....... ( -31.40, z-score = -4.85, R) >droYak2.chr3L 10707878 95 + 24197627 ------CUCUGGGAUAAAA-GCAGCGAACAUU-----UUUGGGGUGUGGGUUCGAGUGUAAAUAAAACCCAUUUAAAAAAUUACGCUGUUUUUAUAAGGUAACAAUG ------.(((...((((((-((((((...(((-----(((.....(((((((..(.......)..)))))))...))))))..)))))))))))).)))........ ( -26.00, z-score = -3.23, R) >droEre2.scaffold_4784 10696457 93 + 25762168 ------CUCUGGGAUAAAA-GCAGUGAACAUU-----UU--GGGUGUGGGUUCGAGUGUAAAUAAAACCCAUUUAAAAAAUUACGCUGUUUUUAUAAGGUAACAAUG ------.(((...((((((-((((((...(((-----((--....(((((((..(.......)..)))))))....)))))..)))))))))))).)))........ ( -24.20, z-score = -2.97, R) >droAna3.scaffold_13337 15609316 106 + 23293914 CCGGGGAUAUGGGAUAAAA-GCGGCGAGAGCUGAAAAUUCACAGAACGGGAAAGUGUGUAAAUAAAACCCAUUUAAAAAAUUACGCUGUUUUUAUAAGGUAACAAUG (((......))).((((((-((((((.(..(((........)))..)(((....(((....)))...))).............))))))))))))............ ( -22.60, z-score = -1.68, R) >consensus _____GCUCUGGGAUAAAA_GCAGCGAACAUU_____UU__GGGUGUGGGUUCGAGUGUAAAUAAAACCCAUUUAAAAAAUUACGCUGUUUUUAUAAGGUAACAAUG ......((((...((((((.((((((.(((((..........)))))(((((...((....))..))))).............)))))))))))).))))....... (-16.04 = -16.36 + 0.32)

| Location | 10,697,592 – 10,697,689 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.12 |
| Shannon entropy | 0.28982 |
| G+C content | 0.35887 |
| Mean single sequence MFE | -18.26 |
| Consensus MFE | -8.66 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
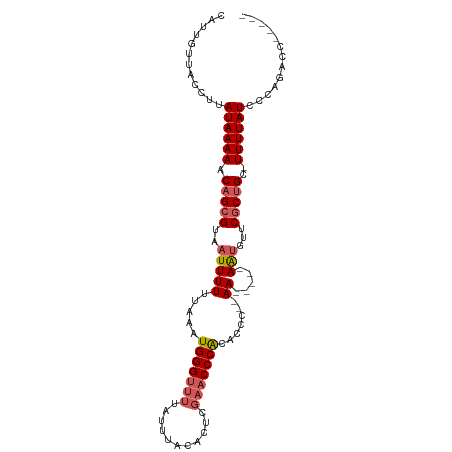
>dm3.chr3L 10697592 97 - 24543557 CAUUGUUACCUUAUAAAAACAGCGUAAUUUUUUAAAUGGGUUUUAUUUACACUCGAACCCACACCC--AA-----AAUGUACGCUGCUUUUUAUACCACGCCAG--- ...........((((((((((((((((((((.....(((((((...........))))))).....--))-----))).))))))).)))))))).........--- ( -23.00, z-score = -4.55, R) >droSec1.super_0 2918363 96 - 21120651 CAUUGUUACCUUAUAAAAACAGCGUAAUUUUUUAAAUGGGUUUUAUUUACACUCGAACCCACACCA--AA-----AAUGUUCGCUGC-UUUUAUACGAGGCCAG--- ........((((((((((.(((((..((((((....(((((((...........)))))))....)--))-----)))...))))).-))))))..))))....--- ( -24.20, z-score = -4.06, R) >droYak2.chr3L 10707878 95 - 24197627 CAUUGUUACCUUAUAAAAACAGCGUAAUUUUUUAAAUGGGUUUUAUUUACACUCGAACCCACACCCCAAA-----AAUGUUCGCUGC-UUUUAUCCCAGAG------ ............((((((.(((((..((((((....(((((((...........)))))))......)))-----)))...))))).-)))))).......------ ( -19.00, z-score = -3.26, R) >droEre2.scaffold_4784 10696457 93 - 25762168 CAUUGUUACCUUAUAAAAACAGCGUAAUUUUUUAAAUGGGUUUUAUUUACACUCGAACCCACACCC--AA-----AAUGUUCACUGC-UUUUAUCCCAGAG------ ............((((((.(((.(..(((((.....(((((((...........))))))).....--))-----)))...).))).-)))))).......------ ( -13.70, z-score = -1.48, R) >droAna3.scaffold_13337 15609316 106 - 23293914 CAUUGUUACCUUAUAAAAACAGCGUAAUUUUUUAAAUGGGUUUUAUUUACACACUUUCCCGUUCUGUGAAUUUUCAGCUCUCGCCGC-UUUUAUCCCAUAUCCCCGG ....(((((((.........)).))))).......(((((....(((((((.((......))..)))))))....(((.......))-).....)))))........ ( -11.40, z-score = 0.42, R) >consensus CAUUGUUACCUUAUAAAAACAGCGUAAUUUUUUAAAUGGGUUUUAUUUACACUCGAACCCACACCC__AA_____AAUGUUCGCUGC_UUUUAUCCCAGACC_____ ............(((((..((((((((....)))..(((((((...........)))))))....................)))))...)))))............. ( -8.66 = -9.50 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:50 2011