| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,678,146 – 10,678,239 |
| Length | 93 |
| Max. P | 0.990342 |

| Location | 10,678,146 – 10,678,239 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Shannon entropy | 0.45577 |
| G+C content | 0.59378 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
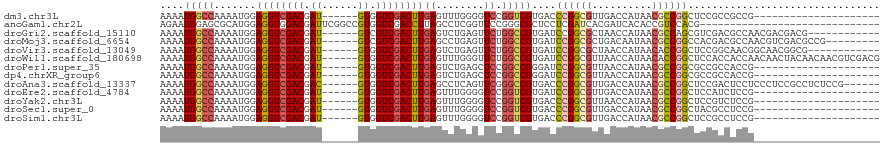
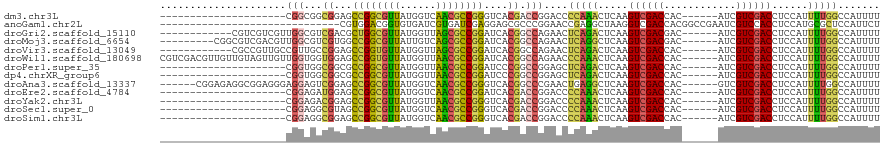
>dm3.chr3L 10678146 93 + 24543557 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUUUGGGGUCCGGUCGUGACCCGGCGUUGACCAUAACGCCGGCUCCGCCGCCG--------------------- .....((((.((((..((((((((...------...))))))))..))))..))))(((.((.((.(((((((((....))))))))).)))))))...--------------------- ( -41.80, z-score = -2.29, R) >anoGam1.chr2L 17354593 90 + 48795086 AGAAUGGAGCGCAUGGAGGUGGACGAUUCGGCCGUGGUCGACCUUAGCCUCGGUUCCGGGCGCUCCUCGAUCACGAUCACACCGUCCACG------------------------------ ..................(((((((....(..(((((((((....(((((((....)))).)))..)))))))))..)....))))))).------------------------------ ( -36.30, z-score = -0.89, R) >droGri2.scaffold_15110 2784673 102 + 24565398 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUCGUCGACUUGAGUCUGAGUUCUGGCCGUGAUCCGGCGCUAACCAUAACGCCAGCGUCGACGCCAACGACGACG------------ ...(((((((.((((((((((((((..------..))))))))....)))..))).))))))).....((((.((....)).))))..(((((.((....)).)))))------------ ( -37.40, z-score = -1.80, R) >droMoj3.scaffold_6654 440964 105 - 2564135 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUCGUCGACUUGAGCCUGAGUUCUGGCCGUGAUCCGGCGCUGACAAUAACGCCGGCCACGACGCCAACGUCGACGCCG--------- ...(((((((......(((((((((..------..)))))))))((((....))))))))))).....((((..........))))(((..(((((....)))))..))).--------- ( -44.20, z-score = -2.92, R) >droVir3.scaffold_13049 1696650 102 + 25233164 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUCUGAGUUCUGGCCGUGAUCCGGCGCUAACCAUAACACCGGCUCCGGCAACGGCAACGGCG------------ ......(((.......((((((((...------...))))))))..............(((((...((((.(((............))).))))..)))))...))).------------ ( -36.10, z-score = -0.93, R) >droWil1.scaffold_180698 6235712 114 + 11422946 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUUUGGGUUCUGGCCGUGAUCCGGCGUUAACCAUAACACCGGCUCCACCACCAACAACUACAACAACGUCGACG .......((.....))..(((((((.(------(((((((((((......)))))..))))(((..((((.((((....)))).))))...)))..............))).))))))). ( -34.30, z-score = -0.51, R) >droPer1.super_35 367979 93 - 973955 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUCUGAGCUCCGGCCGGGAUCCGGCGUUAACCAUAACGCCGGCGCCGCCACCG--------------------- ....(((((.......((((((((...------...))))))))((((....)))).)))))((..(((((((((....)))))))))..)).......--------------------- ( -40.90, z-score = -2.54, R) >dp4.chrXR_group6 9117545 93 + 13314419 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUCUGAGCUCCGGCCGGGAUCCGGCGUUAACCAUAACGCCGGCGCCGCCACCG--------------------- ....(((((.......((((((((...------...))))))))((((....)))).)))))((..(((((((((....)))))))))..)).......--------------------- ( -40.90, z-score = -2.54, R) >droAna3.scaffold_13337 15590133 108 + 23293914 AAAAUGGCCAAAAUGGAGGUCGACGAC------GUGGUCGACUUGAGCCUCAGUUCGGGCCGUGACCCGGCGUUGACCAUAACGCCGGCUCCGACUCCUCCCUCCGCCUCUCCG------ .....(((......((((((((((...------...))))))..(((((.......(((......)))(((((((....))))))))))))...)))).......)))......------ ( -41.92, z-score = -1.89, R) >droEre2.scaffold_4784 10676854 93 + 25762168 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUUUGGGGUCCGGUCGUGAUCCGGCGUUGACCAUAACGCCGGCUCCAUCUCCG--------------------- .....((((.((((..((((((((...------...))))))))..))))..))))(((..(((..(((((((((....)))))))))...)))..)))--------------------- ( -38.70, z-score = -2.31, R) >droYak2.chr3L 10676964 93 + 24197627 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUUUGGGGUCCGGUCGUGACCCGGCGUUAACCAUAACGCCGGCUCCGUCUCCG--------------------- .....((((.((((..((((((((...------...))))))))..))))..))))(((.((.((.(((((((((....))))))))).))))...)))--------------------- ( -41.30, z-score = -2.92, R) >droSec1.super_0 2899467 93 + 21120651 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUUUGGGGUCCGGUCGUGACCCGGCGUUGACCAUAACGCCGGCUACGCCUCCG--------------------- .....((((.((((..((((((((...------...))))))))..))))..))))(((..(((..(((((((((....)))))))))...)))..)))--------------------- ( -40.30, z-score = -2.42, R) >droSim1.chr3L 10069900 93 + 22553184 AAAAUGGCCAAAAUGGAGGUCGACGAU------GUGGUCGACUUGAGUUUGGGGUCCGGUCGUGACCCGGCGUUGACCAUAACGCCGGCUCCGCCUCCG--------------------- .....((((.((((..((((((((...------...))))))))..))))..))))(((..(((..(((((((((....)))))))))...)))..)))--------------------- ( -40.30, z-score = -2.17, R) >consensus AAAAUGGCCAAAAUGGAGGUCGACGAU______GUGGUCGACUUGAGUCUGGGUUCCGGCCGUGAUCCGGCGUUGACCAUAACGCCGGCUCCGCCUCCG_____________________ ....(((((.......((((((((.((......)).))))))))((........)).)))))....((((((..........))))))................................ (-24.38 = -24.46 + 0.08)
| Location | 10,678,146 – 10,678,239 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.22 |
| Shannon entropy | 0.45577 |
| G+C content | 0.59378 |
| Mean single sequence MFE | -40.01 |
| Consensus MFE | -22.55 |
| Energy contribution | -21.80 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
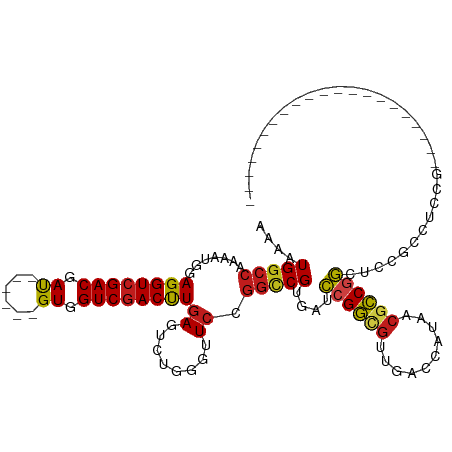
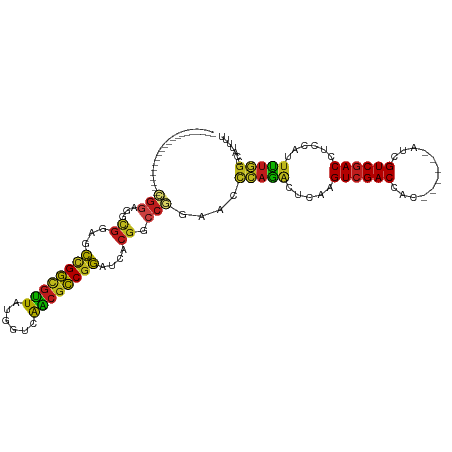
>dm3.chr3L 10678146 93 - 24543557 ---------------------CGGCGGCGGAGCCGGCGUUAUGGUCAACGCCGGGUCACGACCGGACCCCAAACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ---------------------.(((((((((.((((((((......)))))))).)).)).))......((((.....((((((...------...))))))......)))))))..... ( -37.60, z-score = -2.30, R) >anoGam1.chr2L 17354593 90 - 48795086 ------------------------------CGUGGACGGUGUGAUCGUGAUCGAGGAGCGCCCGGAACCGAGGCUAAGGUCGACCACGGCCGAAUCGUCCACCUCCAUGCGCUCCAUUCU ------------------------------.(((((((((.((..((((.((((.....(((((....)).))).....)))).))))..)).))))))))).................. ( -35.40, z-score = -0.90, R) >droGri2.scaffold_15110 2784673 102 - 24565398 ------------CGUCGUCGUUGGCGUCGACGCUGGCGUUAUGGUUAGCGCCGGAUCACGGCCAGAACUCAGACUCAAGUCGACGAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ------------(((((.((....)).)))))(((((((((....))))))))).....((((((((...((......(((((((..------..)))))))))...))))))))..... ( -45.00, z-score = -3.96, R) >droMoj3.scaffold_6654 440964 105 + 2564135 ---------CGGCGUCGACGUUGGCGUCGUGGCCGGCGUUAUUGUCAGCGCCGGAUCACGGCCAGAACUCAGGCUCAAGUCGACGAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ---------.(((((((....)))))))((((((((((((......)))))))).))))((((((((....((.....(((((((..------..)))))))..)).))))))))..... ( -50.80, z-score = -4.38, R) >droVir3.scaffold_13049 1696650 102 - 25233164 ------------CGCCGUUGCCGUUGCCGGAGCCGGUGUUAUGGUUAGCGCCGGAUCACGGCCAGAACUCAGACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ------------.((((((...(((((((((.(((((((((....))))))))).)).))))...)))...)))....((((((...------...)))))).........)))...... ( -38.50, z-score = -2.57, R) >droWil1.scaffold_180698 6235712 114 - 11422946 CGUCGACGUUGUUGUAGUUGUUGGUGGUGGAGCCGGUGUUAUGGUUAACGCCGGAUCACGGCCAGAACCCAAACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU .((((((...(((...(((.(((((.((((..(((((((((....))))))))).)))).))))))))...)))....))))))...------...(((((........)))))...... ( -42.00, z-score = -2.18, R) >droPer1.super_35 367979 93 + 973955 ---------------------CGGUGGCGGCGCCGGCGUUAUGGUUAACGCCGGAUCCCGGCCGGAGCUCAGACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ---------------------.((((((((..(((((((((....)))))))))...)).)))((((.((.(((....)))(((...------...))))).))))......)))..... ( -40.80, z-score = -2.73, R) >dp4.chrXR_group6 9117545 93 - 13314419 ---------------------CGGUGGCGGCGCCGGCGUUAUGGUUAACGCCGGAUCCCGGCCGGAGCUCAGACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ---------------------.((((((((..(((((((((....)))))))))...)).)))((((.((.(((....)))(((...------...))))).))))......)))..... ( -40.80, z-score = -2.73, R) >droAna3.scaffold_13337 15590133 108 - 23293914 ------CGGAGAGGCGGAGGGAGGAGUCGGAGCCGGCGUUAUGGUCAACGCCGGGUCACGGCCCGAACUGAGGCUCAAGUCGACCAC------GUCGUCGACCUCCAUUUUGGCCAUUUU ------......(((.((((..((((...(((((((((((......))))))((((....)))).......)))))..((((((...------...)))))))))).)))).)))..... ( -48.90, z-score = -2.20, R) >droEre2.scaffold_4784 10676854 93 - 25762168 ---------------------CGGAGAUGGAGCCGGCGUUAUGGUCAACGCCGGAUCACGACCGGACCCCAAACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ---------------------(.(((((((((((((((((......))))))))........................((((((...------...))))))))))))))).)....... ( -33.90, z-score = -2.33, R) >droYak2.chr3L 10676964 93 - 24197627 ---------------------CGGAGACGGAGCCGGCGUUAUGGUUAACGCCGGGUCACGACCGGACCCCAAACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ---------------------.((((...(((..(((((((....)))))))(((((.(....))))))....)))..((((((...------...)))))))))).............. ( -36.40, z-score = -2.67, R) >droSec1.super_0 2899467 93 - 21120651 ---------------------CGGAGGCGUAGCCGGCGUUAUGGUCAACGCCGGGUCACGACCGGACCCCAAACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ---------------------.((.((((((.((((((((......)))))))).).))).)).....(((((.....((((((...------...))))))......)))))))..... ( -33.90, z-score = -1.81, R) >droSim1.chr3L 10069900 93 - 22553184 ---------------------CGGAGGCGGAGCCGGCGUUAUGGUCAACGCCGGGUCACGACCGGACCCCAAACUCAAGUCGACCAC------AUCGUCGACCUCCAUUUUGGCCAUUUU ---------------------.((.((((((.((((((((......)))))))).)).)).)).....(((((.....((((((...------...))))))......)))))))..... ( -36.10, z-score = -2.04, R) >consensus _____________________CGGAGGCGGAGCCGGCGUUAUGGUCAACGCCGGAUCACGGCCGGAACCCAGACUCAAGUCGACCAC______AUCGUCGACCUCCAUUUUGGCCAUUUU .....................(((...((...((((((((......))))))))....)).)))....(((((.....((((((............))))))......)))))....... (-22.55 = -21.80 + -0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:45 2011