| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,675,554 – 10,675,615 |
| Length | 61 |
| Max. P | 0.728463 |

| Location | 10,675,554 – 10,675,615 |
|---|---|
| Length | 61 |
| Sequences | 12 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 69.61 |
| Shannon entropy | 0.60798 |
| G+C content | 0.36695 |
| Mean single sequence MFE | -10.47 |
| Consensus MFE | -4.00 |
| Energy contribution | -4.02 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728463 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 10675554 61 + 24543557 CAAACAGAGCAACUCACAAACGCUAAUU-AAGCACAUUUAGCGCGCUAUUAAGCACA-AUAAU------ ......(((...))).....((((((..-........)))))).(((....)))...-.....------ ( -8.70, z-score = -1.34, R) >droSim1.chr3L 10067332 61 + 22553184 CAAACAGAGCAACUCACAAACGCUAAUU-AAGCACAUUUAGCGCGCUAUUAAGCACA-AUAAU------ ......(((...))).....((((((..-........)))))).(((....)))...-.....------ ( -8.70, z-score = -1.34, R) >droSec1.super_0 2896365 61 + 21120651 CAAACAGAGCAACUCACAAACGCUAAUU-AAGCACAUUUAGCGCGCUAUUAAGCACA-AUAAU------ ......(((...))).....((((((..-........)))))).(((....)))...-.....------ ( -8.70, z-score = -1.34, R) >droYak2.chr3L 10674339 61 + 24197627 CAAACUGAGCAACUCACAAAUGCUAAUU-AAGCACAUUUAGCGCGCUAUUAAGCACA-AUAAU------ .....((((...)))).....(((((..-........)))))(.(((....))).).-.....------ ( -7.80, z-score = 0.07, R) >droEre2.scaffold_4784 10674136 61 + 25762168 CAAACUGAGCAGCUCACAAAUGCUAAUU-AAGCACAUUUAGCGCGCUAUUAAGCACA-AUAGU------ ...((((.((.(((......((((....-.)))).....)))))(((....)))...-.))))------ ( -10.90, z-score = -0.26, R) >droAna3.scaffold_13337 15587932 63 + 23293914 -----CAAAGAACGC-UGCGGAAUAAAUGAAUAAAAGCAAACUGGUCACUAAUUGUGGAAUAUUUAGCA -----........((-(((.................)).......((((.....)))).......))). ( -6.23, z-score = 0.67, R) >dp4.chrXR_group6 9114980 54 + 13314419 -------CAACGCCC-UGGGCGCUAAUUGAAUCCCAUUUAGCGCUCUAUUAAACACA-AUUAU------ -------........-.(((((((((.((.....)).)))))))))...........-.....------ ( -15.10, z-score = -3.75, R) >droPer1.super_35 365424 54 - 973955 -------CAACGCCC-UGGGCGCUAAUUGAAUCCCAUUUAGCGCUCUAUUAAACACA-AUUAU------ -------........-.(((((((((.((.....)).)))))))))...........-.....------ ( -15.10, z-score = -3.75, R) >droWil1.scaffold_180698 6232839 56 + 11422946 -----CUAAAAGCUU-UGAAUGCUAAUUGAAAUACAUUUAGCGCACUAUUAAACACA-AUUAU------ -----..........-.....(((((.((.....)).)))))...............-.....------ ( -5.80, z-score = -0.18, R) >droVir3.scaffold_13049 1694344 61 + 25233164 CACACAGCUAAGCUU-AAAGCGCUAAUUGAAACACAUUUAGCGCGCUAUUAAACACA-AUUAU------ ..........(((..-...(((((((.((.....)).))))))))))..........-.....------ ( -11.90, z-score = -2.16, R) >droMoj3.scaffold_6654 438947 61 - 2564135 CACAGAGCUUAACUU-UAAGCGCUAAUUGAAACACAUUUAGCGCGCUAUUAAACACA-AUUAU------ .....((((((....-)))(((((((.((.....)).))))))))))..........-.....------ ( -12.10, z-score = -2.41, R) >droGri2.scaffold_15110 2782303 61 + 24565398 GACACAGUUUAGCUU-UAAGCGCUAAUUGAAACACAUUUAGCGCGCUAUUAAACACA-AUUAU------ ......((((((...-...(((((((.((.....)).)))))))....))))))...-.....------ ( -14.60, z-score = -3.32, R) >consensus CAAACAGAGCAACUC_UAAACGCUAAUUGAAGCACAUUUAGCGCGCUAUUAAACACA_AUUAU______ ....................((((((...........)))))).......................... ( -4.00 = -4.02 + 0.01)

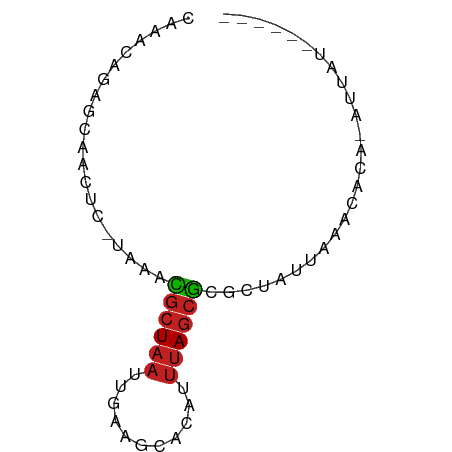
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:42 2011