
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,783,926 – 4,784,023 |
| Length | 97 |
| Max. P | 0.628822 |

| Location | 4,783,926 – 4,784,023 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.85 |
| Shannon entropy | 0.22748 |
| G+C content | 0.36925 |
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -13.04 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
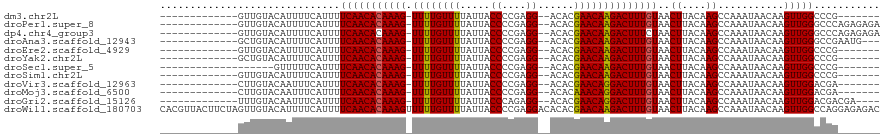
>dm3.chr2L 4783926 97 - 23011544 -------------GUUGUACAUUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGCCCG------- -------------((((..............))))((((((-((((((((.....((....))--....)))))))))))))).........(((((........)))))...------- ( -22.24, z-score = -3.31, R) >droPer1.super_8 1701624 104 + 3966273 -------------GUUGUACAUUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGGCCCAGAGAGA -------------..............(((((...((((((-((((((((.....((....))--....)))))))))))))).........(((.(((.....))).)))...))))). ( -21.90, z-score = -2.02, R) >dp4.chr4_group3 654866 104 + 11692001 -------------GUUGUACAUUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAAGACUUUCUAACUUACAAGCCAAAUAACAAGUUGGGCCCAGAGAGA -------------..............(((((.....((((-((((((((.....((....))--....))))))))))))(((((((..............))))))).....))))). ( -17.24, z-score = -0.66, R) >droAna3.scaffold_12943 1984228 101 + 5039921 -------------GCUGUACAUUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGGCCGAAUG--- -------------......(((((...........((((((-((((((((.....((....))--....)))))))))))))).........(((.(((.....))).))).)))))--- ( -20.30, z-score = -1.73, R) >droEre2.scaffold_4929 4859705 97 - 26641161 -------------GUUGUACAUUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGCCCG------- -------------((((..............))))((((((-((((((((.....((....))--....)))))))))))))).........(((((........)))))...------- ( -22.24, z-score = -3.31, R) >droYak2.chr2L 4792804 97 - 22324452 -------------GCUGUACAUUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGCCCG------- -------------......................((((((-((((((((.....((....))--....)))))))))))))).........(((((........)))))...------- ( -20.70, z-score = -2.85, R) >droSec1.super_5 2880208 91 - 5866729 -------------------GUUUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGCCCG------- -------------------................((((((-((((((((.....((....))--....)))))))))))))).........(((((........)))))...------- ( -20.70, z-score = -3.44, R) >droSim1.chr2L 4653299 97 - 22036055 -------------GUUGUACAUUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGCCCG------- -------------((((..............))))((((((-((((((((.....((....))--....)))))))))))))).........(((((........)))))...------- ( -22.24, z-score = -3.31, R) >droVir3.scaffold_12963 8799782 97 + 20206255 -------------CUUGUACAAUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACGAACAGGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGACGA------- -------------((((((.((......)).....((((((-((((((((.....((....))--....))))))))))))))....))))))((((........))))....------- ( -18.30, z-score = -1.39, R) >droMoj3.scaffold_6500 12193908 97 + 32352404 -------------CUUGUACAAUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCCGAGG--ACACAAACAGGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGACGA------- -------------((((((.((......)).....((((((-((((((((.....((....))--....))))))))))))))....))))))((((........))))....------- ( -17.90, z-score = -1.35, R) >droGri2.scaffold_15126 3334935 100 + 8399593 -------------UUUGUACAAUUUCAUUUUCAACACAAAG-UUUUGUUUUAUUACCCAGAGG--ACACGAACAGGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGACGACGA---- -------------.((((.................((((((-((((((((.....((....))--....))))))))))))))..........((((........))))))))...---- ( -18.30, z-score = -1.39, R) >droWil1.scaffold_180703 1532382 120 + 3946847 CACGUUACUUCUAGUUGUACAUUUUCAUUUUCAACACAAAGUUUUUGUUUUAUUACCCCGAGGACACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGCCAGGAGAGAC ...(((.(((((.((((..............))))((((((((((.((((.....((....))......)))))))))))))).........(((((........))))).))))).))) ( -23.64, z-score = -1.63, R) >consensus _____________GUUGUACAUUUUCAUUUUCAACACAAAG_UUUUGUUUUAUUACCCCGAGG__ACACGAACAAGACUUUGUAACUUACAAGCCAAAUAACAAGUUGGCCCG_______ ..............................(((((((((((.((((((((.....((....))......))))))))))))))..((....))...........)))))........... (-13.04 = -12.86 + -0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:24 2011