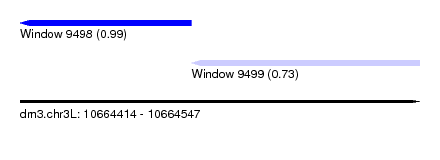
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,664,414 – 10,664,547 |
| Length | 133 |
| Max. P | 0.988161 |

| Location | 10,664,414 – 10,664,471 |
|---|---|
| Length | 57 |
| Sequences | 3 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 97.65 |
| Shannon entropy | 0.03222 |
| G+C content | 0.49405 |
| Mean single sequence MFE | -7.77 |
| Consensus MFE | -7.87 |
| Energy contribution | -7.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10664414 57 - 24543557 CCCCGAAUCACCCGAAUCAACGAGAAACGAAUAAACCAUAUUCGCUUCGACCUGCAG ....((.((....)).)).....(((.((((((.....)))))).)))......... ( -7.90, z-score = -2.45, R) >droSim1.chrU 15391370 55 + 15797150 -CCCGAAUCACCCGAAUCAACGAG-AACGAAUAAACCAUAUUCGCUUCGACCUGCAG -...((.((....)).))..((((-..((((((.....)))))).))))........ ( -7.70, z-score = -2.14, R) >droSec1.super_0 2885492 56 - 21120651 CCCCGAAUCACCCGAAUCAACGAG-AACGAAUAAACCAUAUUCGCUUCGACCUGCAG ....((.((....)).))..((((-..((((((.....)))))).))))........ ( -7.70, z-score = -2.21, R) >consensus CCCCGAAUCACCCGAAUCAACGAG_AACGAAUAAACCAUAUUCGCUUCGACCUGCAG ....((.((....)).))..((((...((((((.....)))))).))))........ ( -7.87 = -7.87 + -0.00)

| Location | 10,664,471 – 10,664,547 |
|---|---|
| Length | 76 |
| Sequences | 7 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.57327 |
| G+C content | 0.40438 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -6.49 |
| Energy contribution | -6.95 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
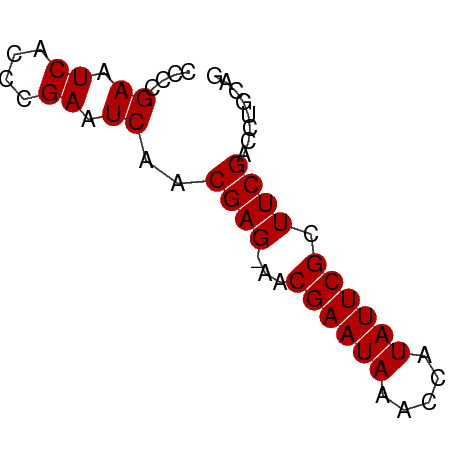
>dm3.chr3L 10664471 76 - 24543557 AGCAUAGCAGCAUCAACAUAUUCGGAUUUUCGGCAUUAC-AAUUUGUGGAAAUUUGCAUA-UUUUGAGGGUGCUGUGA--- ..(((((((.(.((((.((((.((((((((((.((....-....)))))))))))).)))-).)))).).))))))).--- ( -26.00, z-score = -4.28, R) >droSec1.super_0 2885548 79 - 21120651 AGCAGAGCAGCAUCAACAAAUUCGGAUUUUCGGCAUUAC-AAUUUGUGGAAAUUUGCAUA-UUUGGGGGGUGGUGCUGUGA ..((.((((.((((..(((((.((((((((((.((....-....))))))))))))...)-))))...)))).)))).)). ( -26.20, z-score = -3.66, R) >droYak2.chr3L 10663137 75 - 24197627 AUAUGAGCAGCAUCAACCAAUUCGGAUUUUCGGCAUUACAAAUUUGUGGAAAUUUGCAUA-UUUUGGGGGUGCUGA----- .......(((((((..((((..((((((((((.((.........))))))))))))....-..)))).))))))).----- ( -20.00, z-score = -1.87, R) >droEre2.scaffold_4784 10662926 75 - 25762168 AUACGAGCAGCAUCAACCAAUUCGGAUUUUCGGCAUUAC-AAUUUGUGGAAAUUUGCAUAUUUUUGGGGGUGCUGA----- .......(((((((..((((..((((((((((.((....-....)))))))))))).......)))).))))))).----- ( -20.20, z-score = -2.20, R) >droAna3.scaffold_13337 15578146 68 - 23293914 ----GUGUGGCAUCAACAAAUUUCGGAUUACAAUUUUG--A----GGGAAAAUUUGCAUAACAGGGGGGGUGCUAGGA--- ----.(.(((((((..(...(((((((.......))))--)----)).....((((.....)))))..))))))).).--- ( -12.90, z-score = -1.22, R) >dp4.chrXR_group6 9105062 72 - 13314419 -GGCAUGUGCCAUCGACAAAUUCGAAUAUUCGGAUUUAC-AAUUUGGGAAAAUUUGCAU--UUCGGGUUCGAGUGU----- -(((....))).((((.....))))((((((((((((..-....((.((....)).)).--...))))))))))))----- ( -16.10, z-score = -0.55, R) >droPer1.super_35 354620 72 + 973955 -GGCAUGUGCCAUCGACAAAUUCGAAUAUUCGGAUUUAC-AAUUUGGGAAAAUUUGCAU--UUCGGGUUCGAGUGU----- -(((....))).((((.....))))((((((((((((..-....((.((....)).)).--...))))))))))))----- ( -16.10, z-score = -0.55, R) >consensus AGACGAGCAGCAUCAACAAAUUCGGAUUUUCGGCAUUAC_AAUUUGUGGAAAUUUGCAUA_UUUGGGGGGUGCUGU_____ .......(((((((..(..((.(((((((((................))))))))).))......)..)))))))...... ( -6.49 = -6.95 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:40 2011