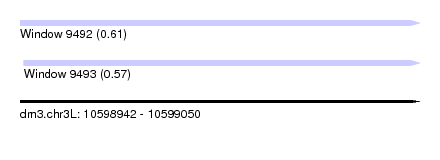
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,598,942 – 10,599,050 |
| Length | 108 |
| Max. P | 0.613446 |

| Location | 10,598,942 – 10,599,050 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Shannon entropy | 0.37974 |
| G+C content | 0.56007 |
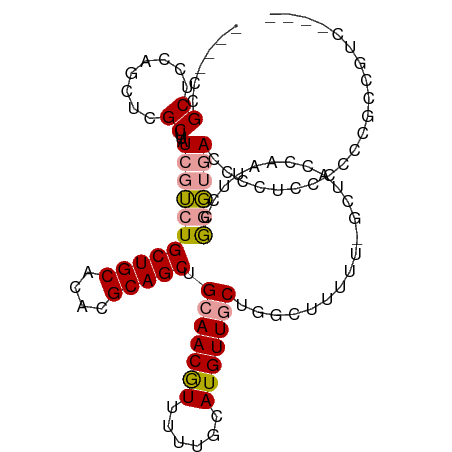
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.23 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
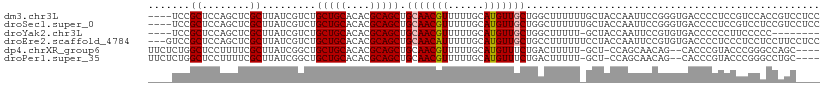
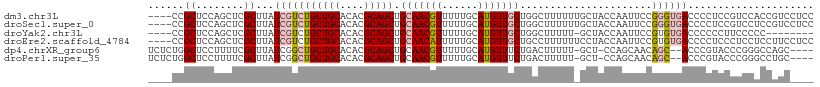
>dm3.chr3L 10598942 108 + 24543557 ----UCCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUUUGCUACCAAUUCCGGGUGACCCCUCCGUCCACCGUCCUCC ----.......((((..((.....))..)))).....((((((((((.....))))).)))))((((......)))).......(((..(((......)))..)))...... ( -26.40, z-score = -0.96, R) >droSec1.super_0 2817268 108 + 21120651 ----UCCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUUUGCUACCAAUUCCGGGUGACCCCUCCGUCCUCCGUCCUCC ----.......((((..((.....))..)))).....((((((((((.....))))).)))))((((......)))).......((((.(((......))).))))...... ( -27.20, z-score = -1.47, R) >droYak2.chr3L 10595164 99 + 24197627 ----UCCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUU-GCUACCAAUUCCGUGUGACCCCCCUUCCCCC-------- ----.......((((..((.....))..))))(((((((((((((((.....))))).)))))((((.....-))))........)))))..............-------- ( -27.20, z-score = -3.22, R) >droEre2.scaffold_4784 10595461 109 + 25762168 ---GUCCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACAUUUUUGCAUGUUGCUGCCUUUUUUCCUACCAAUUCCGUGUGACCCCUCCCUCCUCCUUCCUCC ---((((((..((((..((.....))..)))).....((((((((((.....))))).)))))......................))).))).................... ( -22.60, z-score = -3.78, R) >dp4.chrXR_group6 9041298 104 + 13314419 UUCUCUGGCUCCUUUUCGCUUAUCGGCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUUCUGACUUUUU-GCU-CCAGCAACAG--CACCCGUACCCGGGCCAGC---- ....(((((((..............((.(((((....))))).)).(((....(((.((((.(((.......-...-.))).)))))--))..)))....))))))).---- ( -31.50, z-score = -1.54, R) >droPer1.super_35 288291 104 - 973955 UUCUCUGGCUCCUUUUCGCUUAUCGGCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUUCUGACUUUUU-GCU-CCAGCAACAG--CACCCGUACCCGGGCCUGC---- ......(((((..............((.(((((....))))).)).(((....(((.((((.(((.......-...-.))).)))))--))..)))....)))))...---- ( -28.00, z-score = -0.51, R) >consensus ____UCCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUU_GCUACCAAUUCCGGGUGACCCCUCCCCCCGCCGUC____ .......((........)).........(((((....))))).(((((((......)))))))................................................. (-14.04 = -14.23 + 0.20)
| Location | 10,598,943 – 10,599,050 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Shannon entropy | 0.37001 |
| G+C content | 0.56475 |
| Mean single sequence MFE | -26.88 |
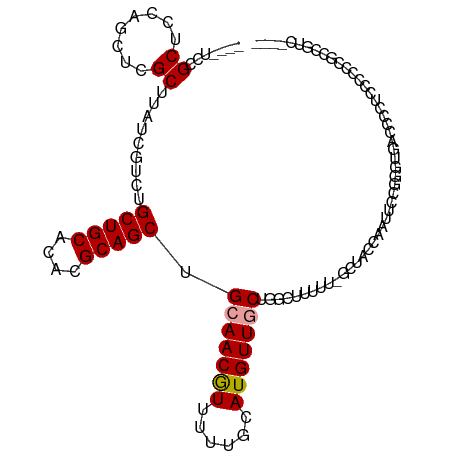
| Consensus MFE | -14.60 |
| Energy contribution | -15.13 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10598943 107 + 24543557 ----CCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUUUGCUACCAAUUCCGGGUGACCCCUCCGUCCACCGUCCUCC ----......((((..((.....))..)))).....((((((((((.....))))).)))))((((......)))).......(((..(((......)))..)))...... ( -26.40, z-score = -0.94, R) >droSec1.super_0 2817269 107 + 21120651 ----CCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUUUGCUACCAAUUCCGGGUGACCCCUCCGUCCUCCGUCCUCC ----......((((..((.....))..)))).....((((((((((.....))))).)))))((((......)))).......((((.(((......))).))))...... ( -27.20, z-score = -1.41, R) >droYak2.chr3L 10595165 98 + 24197627 ----CCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUU-GCUACCAAUUCCGUGUGACCCCCCUUCCCCC-------- ----......((((..((.....))..))))(((((((((((((((.....))))).)))))((((.....-))))........)))))..............-------- ( -27.20, z-score = -3.17, R) >droEre2.scaffold_4784 10595463 107 + 25762168 ----CCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACAUUUUUGCAUGUUGCUGCCUUUUUUCCUACCAAUUCCGUGUGACCCCUCCCUCCUCCUUCCUCC ----......((((..((.....))..))))((((((..((.(((((((......))))))).))..................))))))...................... ( -20.95, z-score = -3.29, R) >dp4.chrXR_group6 9041299 103 + 13314419 UCUCUGGCUCCUUUUCGCUUAUCGGCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUUCUGACUUUUU-GCU-CCAGCAACAGC--ACCCGUACCCGGGCCAGC---- ...(((((((..............((.(((((....))))).)).(((....(((.((((.(((.......-...-.))).))))))--)..)))....))))))).---- ( -31.50, z-score = -1.60, R) >droPer1.super_35 288292 103 - 973955 UCUCUGGCUCCUUUUCGCUUAUCGGCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUUCUGACUUUUU-GCU-CCAGCAACAGC--ACCCGUACCCGGGCCUGC---- .....(((((..............((.(((((....))))).)).(((....(((.((((.(((.......-...-.))).))))))--)..)))....)))))...---- ( -28.00, z-score = -0.55, R) >consensus ____CCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUU_GCUACCAAUUCCGGGUGACCCCUCCCCCCGCCGUC____ ......((........))...(((((((((((....))))).(((((((......)))))))......................))))))..................... (-14.60 = -15.13 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:35 2011