
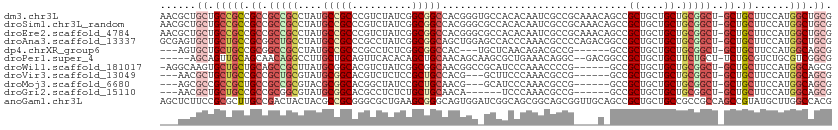
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,578,481 – 10,578,592 |
| Length | 111 |
| Max. P | 0.696920 |

| Location | 10,578,481 – 10,578,592 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.79 |
| Shannon entropy | 0.67084 |
| G+C content | 0.70682 |
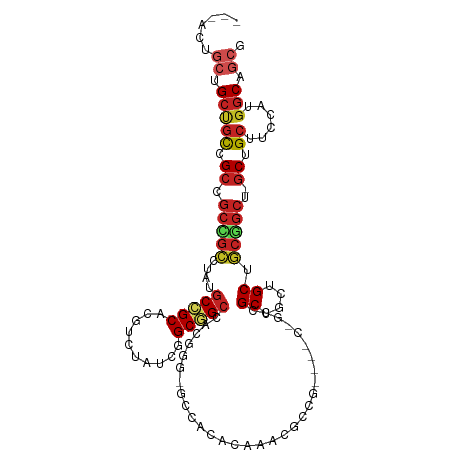
| Mean single sequence MFE | -49.75 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.85 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696920 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 10578481 111 + 24543557 AACGCUGCUGCCGCCGCCGCCGCCUAUGCCGCCCGUCUAUCGGCGGCCACGGGUGCCACACAAUCGCCGCAAACAGCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCUGCG ...((....))(((.((((.((((..(((((((........))))).))..)))).)...........(((..(((((((.......))))))-).)))......))).))) ( -46.20, z-score = 0.28, R) >droSim1.chr3L_random 483379 111 + 1049610 AACGCUGCUGCCGCCGCCGCCGCCUAUGCCGCCCGUCUAUCGGCGGCCACGGGCGCCACACAAUCGCCGCAAACAGCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCUGCG ...((....))(((.((((.((((..(((((((........))))).))..)))).)...........(((..(((((((.......))))))-).)))......))).))) ( -48.10, z-score = -0.06, R) >droEre2.scaffold_4784 10574917 111 + 25762168 AACGCUGCUGCCGCCGCCGCCGCCUAUGCCGCCCGUCUAUCGGCGGCCACGGGCGCCACACAAUCGCCGCAAACAGCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCUGCG ...((....))(((.((((.((((..(((((((........))))).))..)))).)...........(((..(((((((.......))))))-).)))......))).))) ( -48.10, z-score = -0.06, R) >droAna3.scaffold_13337 15485472 111 + 23293914 GCGAGUGCUGCUGCCGCGGCUGCCUAUGCCGCCCGCCUAUCGGCGGCAGCUGGAGCCACCCAAACGCCCCAGACGGCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCUGCG (((((((((((.((.(((((.(((..(((((((........)))))))(((((.((.........)).)))).)))).))))).)).))))).-)))(((.....)))))). ( -53.80, z-score = -0.74, R) >dp4.chrXR_group6 8997874 99 + 13314419 ---AGUGCUGCUGCCGCGGCCGCCUAUGCCGCCCGCCUCUCGGCGGCCAC---UGCUCAACAGACGCCG------GCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCAGCG ---...(((((.((.(((((.(((...((((((........))))))..(---((.....))).....)------)).))))).)).))))).-((((((.....)))))). ( -50.80, z-score = -1.57, R) >droPer1.super_4 6591745 104 - 7162766 -----AGCAGUUGCAGCAACAGGCCUUGCUGCAGUUCACACAGCUGCAACAGCAAGCGCUGAAACAGGC--GACGGCCGCUGCUGCUUCUGCU-UCUGCGUCUGCGUCGGCG -----(((((..((((((...(((((((((((((((.....))))))..((((....)))).....)))--)).))))..))))))..)))))-.(((((....)).))).. ( -46.90, z-score = -0.71, R) >droWil1.scaffold_181017 149279 104 - 939937 -AGGCAAGUGCUGCUGCAGCCGCUUAUGCGGCACGUCUAUCGGCGGCAACGGCCGCAUCCCAAACCCCG------GCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCAGCG -.(((....)))(((((.(((((....))))).........(((((((.((((((............))------)))).)))))))))))).-((((((.....)))))). ( -54.30, z-score = -2.03, R) >droVir3.scaffold_13049 16377191 99 + 25233164 ---AACGCUGCUGCCGCCGCUGCGUAUGCGGCACGUCUCUCCGCUGCCACG---GCUUCCCAAACGCCG------GCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCAGCG ---.(((.(((((((((....)))...))))))))).....((((((((((---((.........))))------((.(((((....))))).-))........)))))))) ( -44.70, z-score = -1.32, R) >droMoj3.scaffold_6680 4415933 99 - 24764193 ---AGCGCCGCCGCUGCCGCCGCGUACGCGGCACGGCUAUCCGCUGCAACG---GCAUCCCAAACGCCG------GCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCAGCG ---((((....))))(((((.(((((.(((((.((((.....))))...((---((.........))))------)))))))).)).))))).-((((((.....)))))). ( -49.50, z-score = -0.63, R) >droGri2.scaffold_15110 12260039 96 - 24565398 ---AACGCUGCUGCCGCCGCGGCGUAUGCGGCACGCCUCUCUGCUGCAACA------UCCCAAACGCCG------GCCGCUGCUGCUGCGGCU-GCUGCUUCCAUGGCAGCG ---...(((((.((.((.(((((((.(((((((........))))))))).------...(.......)------))))).)).)).))))).-((((((.....)))))). ( -44.60, z-score = -1.46, R) >anoGam1.chr3L 29742762 112 - 41284009 AGCUCUUCCGCGCUUGCCGACUACUACGCCGCGGGCGCUGAAGCGGGCAGUGGAUCGGCAGCGGCAGCGGUUGCAGCCGCUGCUGCCGCCGCCAGCCGUAUGCUUGGCCACG .(((((((.((((((((.(.(......)).)))))))).))))..))).((((..((((.(((((((((((....))))))))))).))))(((((.....)).))))))). ( -60.30, z-score = -1.28, R) >consensus ___ACUGCUGCUGCCGCCGCCGCCUAUGCCGCACGUCUAUCGGCGGCCACGGG_GCCACACAAACGCCG____C_GCCGCUGCUGCUGCGGCU_GCUGCUUCCAUGGCAGCG ......(((((.((.((.((.((....(((((..........))))).......(.....)..............)).)).)).)).)))))..((.(((.....))).)). (-22.74 = -23.85 + 1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:31 2011