| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,571,722 – 10,571,813 |
| Length | 91 |
| Max. P | 0.910585 |

| Location | 10,571,722 – 10,571,813 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 62.10 |
| Shannon entropy | 0.76426 |
| G+C content | 0.52019 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -12.35 |
| Energy contribution | -12.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
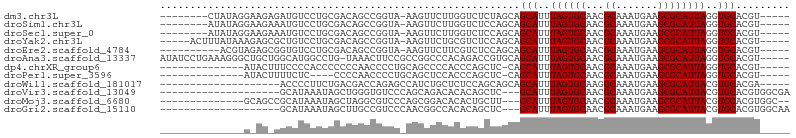
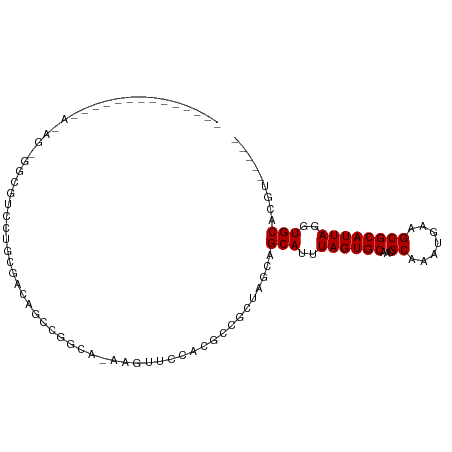
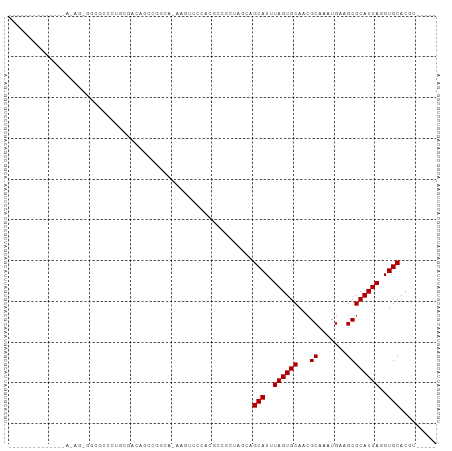
>dm3.chr3L 10571722 91 - 24543557 --------CUAUAGGAAGAGAUGUCCUGCGACAGCCGGUA-AAGUUCUUGGUCUCUAGCAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU----- --------(((.(((..((((...((.((....)).))..-....))))..))).)))..(((((((((((...((.......)))))))))))))....----- ( -25.20, z-score = -0.38, R) >droSim1.chr3L 9969029 91 - 22553184 --------AUAUAGGAAGAAAUGUCCUGCGACAGCCGGUA-AAGUUCUUGGUCUCCAGCAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU----- --------.....(((((((....((.((....)).))..-...)))).....)))....(((((((((((...((.......)))))))))))))....----- ( -23.60, z-score = -0.28, R) >droSec1.super_0 2790135 91 - 21120651 --------AUAUAGGAAGAAAUGUCCUGCGACAGCCGGUA-AAGUUCUUGGUCUCCAGCAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU----- --------.....(((((((....((.((....)).))..-...)))).....)))....(((((((((((...((.......)))))))))))))....----- ( -23.60, z-score = -0.28, R) >droYak2.chr3L 10567131 94 - 24197627 -----ACUUUAUAAAGAGCGCUGUCCUGCGACAGCCGGUA-AAGUUCUGCGUCUCCAGCAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU----- -----(((((((.....(.((((((....))))))).)))-))))..(((.......)))(((((((((((...((.......)))))))))))))....----- ( -29.60, z-score = -0.89, R) >droEre2.scaffold_4784 10568095 89 - 25762168 ----------ACGUAGAGCGGUGUCCUGCGACAGCCGGUA-AAGUUCUUCGUCUCCAGCAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU----- ----------(((.(((((..((.((.((....)).))))-..))))).)))........(((((((((((...((.......)))))))))))))....----- ( -28.00, z-score = -0.84, R) >droAna3.scaffold_13337 15478824 99 - 23293914 AUAUCCUGAAAGGGCUGCUGGCAUGGCCUG-UAAACUUCCGCCGGCCCACAGACCGUGCAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU----- ....(((....)))......((((((.(((-(...((......))...)))).)))))).(((((((((((...((.......)))))))))))))....----- ( -31.00, z-score = 0.40, R) >dp4.chrXR_group6 8990960 85 - 13314419 --------------AUACUUUCCCCACCCCCCCAACCCCUGCAGCCCCACCCAGCUC-CAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU----- --------------.........................((.(((........))).-))(((((((((((...((.......)))))))))))))....----- ( -16.80, z-score = -1.99, R) >droPer1.super_3596 145 81 + 2515 --------------AUACUUUUCUC----CCCCAACCCCUGCAGCUCCACCCAGCUC-CAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU----- --------------...........----..........((.((((......)))).-))(((((((((((...((.......)))))))))))))....----- ( -18.20, z-score = -1.86, R) >droWil1.scaffold_181017 140162 80 + 939937 --------------------ACCCCUUCUGACGACCAGAGCCAUCUGCUCUCCAGCAGCAGCAUUUAGUGCAAGGCAAAUGAAGCGCAUUACGUGCACGA----- --------------------......((((.....))))(((..(((((....)))))..((((...))))..))).......((((.....))))....----- ( -21.30, z-score = -0.76, R) >droVir3.scaffold_13049 16371209 82 - 25233164 --------------------GCAUAAAUAGCUGGGUGUCCCAGCAGACACACAGCUC---GCAUUUAGUGCAACGCAAAUGAAGCGCAUUACGUGCACGUGGCGA --------------------((......(((((.(((((......))))).))))).---((((.((((((...((.......)))))))).)))).....)).. ( -27.60, z-score = -1.17, R) >droMoj3.scaffold_6680 4409955 86 + 24764193 --------------GCAGCCGCAUAAAUAGCUAGGCGUCCCAGCGGACACACUGCUU---GCAUUUAGUGCAACGCAAAUGAAGCGCAUUACGUGCACGUGGC-- --------------...(((((......(((.(((.((((....)))).).)))))(---((((.((((((...((.......)))))))).))))).)))))-- ( -29.10, z-score = -0.64, R) >droGri2.scaffold_15110 12253801 82 + 24565398 --------------------GCAUAAAUAGCUUGCCGUCCCAACGGCCACACAGCUC---GCAUUUAGUGCAACGCAAAUGAAGCGCAUUACGUGCACGUGGCAA --------------------((......((((.(((((....))))).....)))).---((((.((((((...((.......)))))))).)))).....)).. ( -22.60, z-score = 0.09, R) >consensus ______________A_AG_GGCGUCCUGCGACAGCCGGCA_AAGUUCCACGCCGCUAGCAGCAUUUAGUGCAACGCAAAUGAAGCGCAUUAGGUGCACGU_____ ............................................................(((..((((((...((.......))))))))..)))......... (-12.35 = -12.35 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:30 2011