
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,523,223 – 10,523,315 |
| Length | 92 |
| Max. P | 0.647367 |

| Location | 10,523,223 – 10,523,315 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.87 |
| Shannon entropy | 0.57849 |
| G+C content | 0.40720 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -6.83 |
| Energy contribution | -8.91 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10523223 92 + 24543557 ---------UGUAUGUUGUCAAGCUUUGCAAUUUGCCGAGUUCCUUGGGAA--GUGUCGUAAUAAAUUU-GUGACUGAAGACA----AACCAAGAAC-UCGGCAAAUAA ---------((((.(((....)))..))))(((((((((((((.((((...--.((((...........-.........))))----..))))))))-))))))))).. ( -32.15, z-score = -4.81, R) >droSim1.chr3L 9919823 92 + 22553184 ---------UGUAUGUCGUCAAGCUGUGCAAUUUUCCGAGUUCCUUGGGAA--GUGUCGUAAUAAAUUU-GUGACUGAAGACA----AACCAAGAAC-UCGGCAAAUAA ---------((((((.(.....).))))))((((.((((((((.((((...--.((((...........-.........))))----..))))))))-)))).)))).. ( -25.25, z-score = -2.49, R) >droSec1.super_0 2741888 92 + 21120651 ---------UGUAUGUCGUCAAGCUGUGCAAUUUUCCGAGUUCCUUGGGAA--GUGUCGUAAUAAAUUU-GUGACUGAAGACA----AACCAAGAAC-UCGGCAAAUAA ---------((((((.(.....).))))))((((.((((((((.((((...--.((((...........-.........))))----..))))))))-)))).)))).. ( -25.25, z-score = -2.49, R) >droYak2.chr3L 10516931 100 + 24197627 UAUGUAUGUUGCAUGUCGUCAAGCUGUGCAAUUUUCCGAGUUCCUUGGGAA--GUGUCGUAAUAAAUUU-CUGACUGAAGACA----A-CCAAGAAC-UCGGCAAAUAA ..(((((...((.((....)).)).)))))((((.((((((((.((((...--.((((...........-.........))))----.-))))))))-)))).)))).. ( -27.65, z-score = -2.47, R) >droEre2.scaffold_4784 10517741 96 + 25762168 ---------UGUAUGUCGUCAAGCUGUGCAAUUUUCCGAGUUCCUUGGGAA--GUGUCGUAAUAAAUUU-CUGACUGAAGAACUGACAACCAAGAAC-UCGGCAAAUAA ---------((((((.(.....).))))))((((.((((((((.((((...--.(((((........((-((......)))).))))).))))))))-)))).)))).. ( -26.00, z-score = -2.26, R) >droAna3.scaffold_13337 15435015 96 + 23293914 ------UGCCGAGUGU-ACCAAG-UGU-CGAGUACCUAGUUCCUUUUUGAACUAUGUCGUAAUAAAUUUUGUGACUGAAGACGGAGAACUC---GUC-UCGGCAAAUAA ------(((((((...-......-...-(((((.((((((((......)))))).(((((((......))))))).......))...))))---).)-))))))..... ( -25.52, z-score = -2.01, R) >droPer1.super_35 156741 88 - 973955 ------------------AUAUAGUGUACGAUGUACUG--UUUGUCGAGUUCUGUGUCGCAAUAAAUUU-UCAAAUGUCUCUUCGCCCCUAAGGCUCGCCGCCAAGUAA ------------------...(((.(..(((.(.((.(--((((..(((((.(((......))))))))-.)))))))..).)))..)))).(((.....)))...... ( -11.80, z-score = 0.93, R) >consensus _________UGUAUGUCGUCAAGCUGUGCAAUUUUCCGAGUUCCUUGGGAA__GUGUCGUAAUAAAUUU_GUGACUGAAGACA____AACCAAGAAC_UCGGCAAAUAA ..............................((((.((((((((.((((......((((.....................))))......)))))))).)))).)))).. ( -6.83 = -8.91 + 2.09)

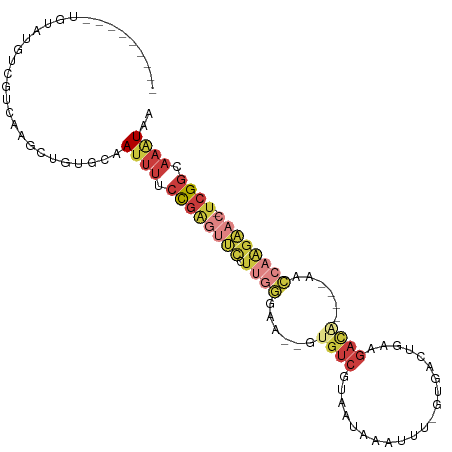
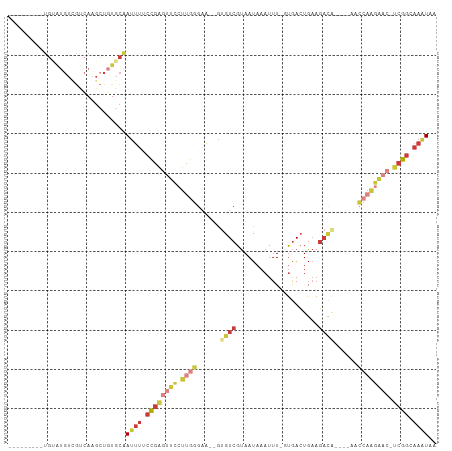
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:27 2011