| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,501,292 – 10,501,390 |
| Length | 98 |
| Max. P | 0.902125 |

| Location | 10,501,292 – 10,501,390 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 63.04 |
| Shannon entropy | 0.72137 |
| G+C content | 0.36477 |
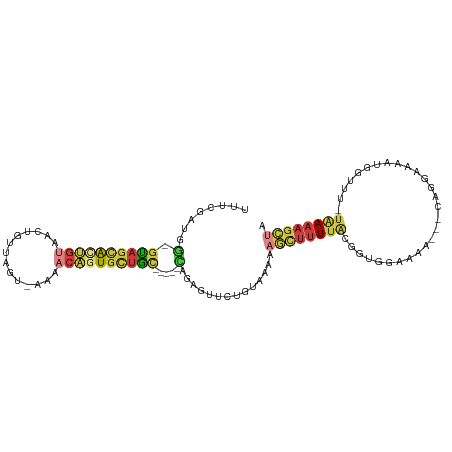
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -8.67 |
| Energy contribution | -9.47 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
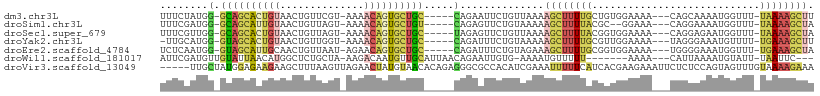
>dm3.chr3L 10501292 98 + 24543557 UUUCUAUGG-GCAGCACUGUAACUGUUCGU-AAAACAGUGCUGC-----CAGAAUUCUGUUAAAAGCUUUUGCUGUGGAAAA---CAGCAAAAUGGUUU-UAAAAGCUU .((((...(-((((((((((.((.....))-...))))))))))-----)))))..((.((((((.((((((((((.....)---)))))))).).)))-))).))... ( -36.40, z-score = -4.84, R) >droSim1.chr3L 9898848 96 + 22553184 UUUCGAUGG-GCAGCAUUGUAACUGUUAGU-AAAACAGUGCUGU-----CAGAGUUCUGUAAAAAGCUUUUACGC--GGAAA---CAGGAAAAUGGUUU-UAAAAGCUA .(((....(-((((((((((.((.....))-...))))))))))-----).)))((((((.....((......))--....)---)))))...(((((.-....))))) ( -23.70, z-score = -1.33, R) >droSec1.super_679 1253 98 + 7707 UUUCGUUGG-GCAGCACUGUAACUGUUAGU-AAAACAGUGCUGC-----UAGAGUUCUGUUAAAAGCUUUUACGGUGGAAAA---CAGGAGAAUGGUUU-UAAAAGCUA .(((....(-((((((((((.((.....))-...))))))))))-----).)))(((((((....(((.....)))....))---)))))...(((((.-....))))) ( -27.30, z-score = -1.61, R) >droYak2.chr3L 10493842 97 + 24197627 -UUGCAUGG-GUAGCACUGUAACUGUUGGU-AAAACAGUGCUGC-----CAGAUUUCUGUAAAAAGCUUUUGCGUUGGAAAA---UAGGGAAAUGUUUU-UGAAAGCUU -((((...(-((((((((((.((.....))-...))))))))))-----)((....)))))).((((((((......(((((---((......))))))-))))))))) ( -28.00, z-score = -2.24, R) >droEre2.scaffold_4784 10495298 98 + 25762168 UCUCAAUGG-GUAGCAUUGCAACUGUUAAU-AGAACAGUGCUGC-----CAGAUUUCUGUAGAAAGCUUUUGCGGUGGAAAA---UGGGGAAAUGGUUU-UGAAAGCUA (((((...(-(((((((((...(((....)-))..)))))))))-----)...((((..(.....((....)).)..)))).---)))))...(((((.-....))))) ( -25.10, z-score = -0.87, R) >droWil1.scaffold_181017 37235 93 - 939937 AUUCGAUGUUGUAUUAACAUGGCUCUGCUA-AAGACAAUGUUGCAUUAACAGAAUUGUG-AAAAUGUUUUU-------AAAA---CAUUAAAAUGUAUU-UAAUUC--- .((..(((((.((..((((((((...))).-...(((((.(((......))).))))).-...)))))..)-------).))---)))..)).......-......--- ( -13.10, z-score = 0.34, R) >droVir3.scaffold_13049 16296449 104 + 25233164 -----UUGCUAUGGAGAAGAAGCUUUAAGUUAGAACUAUGUAACACAGAGGGCGCCACAUCGAAAUUUUUCAUCACGAAGAAAUUCUCUCCAGUAGUUUGUAAAAGAAA -----..((((((((((....(((((..((((.(....).))))....)))))........(((.((((((.....)))))).))))))))).))))............ ( -21.60, z-score = -0.89, R) >consensus UUUCGAUGG_GUAGCACUGUAACUGUUAGU_AAAACAGUGCUGC_____CAGAGUUCUGUAAAAAGCUUUUACGGUGGAAAA___CAGGAAAAUGGUUU_UAAAAGCUA ..........((((((((((..............))))))))))....................((((((((............................)))))))). ( -8.67 = -9.47 + 0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:24 2011