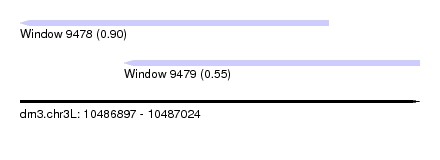
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,486,897 – 10,487,024 |
| Length | 127 |
| Max. P | 0.898808 |

| Location | 10,486,897 – 10,486,995 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Shannon entropy | 0.47911 |
| G+C content | 0.36544 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -10.09 |
| Energy contribution | -10.48 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
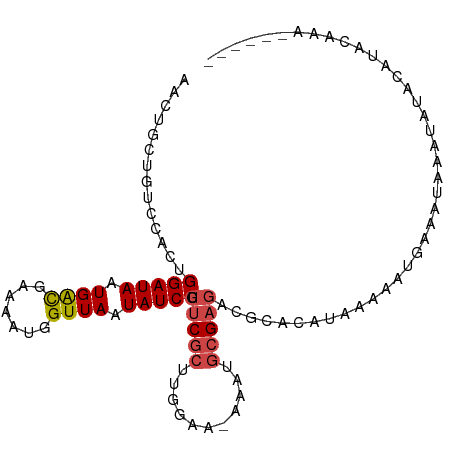
>dm3.chr3L 10486897 98 - 24543557 AACUGCUGUCCACUGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAA-AAAUGCGAGACGCACAUAAAAACGAAAUAAAUAUACAUAUACAAG---- ...(((.(((....(((((.((((.......)))).)))))..((((......-....))))))))))...............................---- ( -17.70, z-score = -1.72, R) >droSim1.chr3L 9884014 96 - 22553184 AACUGCUGUCCACUGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAA-AAAUGCGAGACGCACAUAAAAAUGAAAUAAAUAUACAUACAAG------ ...(((.(((....(((((.((((.......)))).)))))..((((......-....)))))))))).............................------ ( -17.70, z-score = -1.79, R) >droSec1.super_0 2697449 96 - 21120651 AACUGCUGUCCACUGGAUAAUGGCGAAAAUGGUUAAUAUCCGUUCGCUUGGAA-AAAUGCGAGACGCACAUAAAAAUGAAAUAAAUAUACAUACAAG------ ...(((.(((....(((((.((((.......)))).)))))..((((......-....)))))))))).............................------ ( -17.20, z-score = -1.02, R) >droYak2.chr3L 10478635 97 - 24197627 AACUGCUGUCCACAGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAA-AAAAGCGAGCCAUACAUAAAAAUGUAAUAUAUAUACAUACAACC----- ..............(((((.((((.......)))).)))))((((((((....-..))))))))..(((((....)))))..................----- ( -22.10, z-score = -3.03, R) >droEre2.scaffold_4784 10479887 99 - 25762168 AACUGCUGUCCACUGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAAGAAAGGCGAGC----CAUAAAAAUGAAAUAAAUAUACUCGUACAUACAAU ......(((.(...(((((.((((.......)))).)))))((((((((.......))))))))----........................).)))...... ( -19.70, z-score = -1.52, R) >dp4.chrXR_group6 8893295 77 - 13314419 AGGAGCAGCAGCAAGGAUAAUGAUGAAAAUGGUUAAUAUCCGAACG-----AAAAGAGAGGAAACCCA-AGAAACAUUUCGUG-------------------- .(((((....)).....((((.((....)).))))...)))..(((-----(((.....((....)).-.......)))))).-------------------- ( -11.32, z-score = -0.82, R) >consensus AACUGCUGUCCACUGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAA_AAAUGCGAGACGCACAUAAAAAUGAAAUAAAUAUACAUACAAA______ ..............(((((.((((.......)))).))))).(((((...........)))))........................................ (-10.09 = -10.48 + 0.39)

| Location | 10,486,930 – 10,487,024 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 63.48 |
| Shannon entropy | 0.66150 |
| G+C content | 0.45100 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -6.96 |
| Energy contribution | -6.53 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
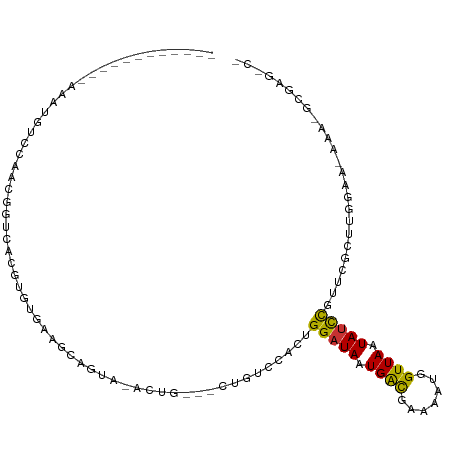
>dm3.chr3L 10486930 94 - 24543557 ------------AAAUGUCCAACGGUCACGUGUGAAGCAGUA-ACUG---CUGUCCACUGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAA-AAAUGCGAGACG ------------.....(((((((..(..(((.(((((((..-.)))---)).))))).(((((.((((.......)))).))))))..)).))))).-............ ( -25.00, z-score = -1.57, R) >droSim1.chr3L 9884045 94 - 22553184 ------------AAAUGUCCAACGGUCACGUGUGAAGCAGUA-ACUG---CUGUCCACUGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAA-AAAUGCGAGACG ------------.....(((((((..(..(((.(((((((..-.)))---)).))))).(((((.((((.......)))).))))))..)).))))).-............ ( -25.00, z-score = -1.57, R) >droSec1.super_0 2697480 94 - 21120651 ------------AAAUGUACAAAGGUCACGUGUGAAGCAGUA-ACUG---CUGUCCACUGGAUAAUGGCGAAAAUGGUUAAUAUCCGUUCGCUUGGAA-AAAUGCGAGACG ------------............(((..(((.(((((((..-.)))---)).))))).(((((.((((.......)))).)))))..((((......-....))))))). ( -24.00, z-score = -1.31, R) >droYak2.chr3L 10478667 94 - 24197627 ------------AAAUGUCCAACGGACACGUGUGAAGCAGUA-ACUG---CUGUCCACAGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAA-AAAAGCGAGCCA ------------...(((((...))))).(((.(((((((..-.)))---)).))))).(((((.((((.......)))).)))))((((((((....-..)))))))).. ( -33.80, z-score = -4.47, R) >droEre2.scaffold_4784 10479920 95 - 25762168 ------------AAAUGUCCAACGGUCACGUGUUAAGCAGUA-ACUG---CUGUCCACUGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAAGAAAGGCGAGCCA ------------...((.((...)).)).(((...(((((..-.)))---))...))).(((((.((((.......)))).)))))((((((((.......)))))))).. ( -27.80, z-score = -1.90, R) >dp4.chrXR_group6 8893317 87 - 13314419 AAAUGGCAUCAGGCACGUGUAAGGGCAACAGCAGG-----------AGCAGCAGCAA--GGAUAAUGAUGAAAAUGGUUAAUAUCCGAACGAAAAGAGAG----------- .....((.....((.(.(((...(....).))).)-----------.))....))..--(((((.((((.......)))).)))))..............----------- ( -15.30, z-score = -0.87, R) >droPer1.super_35 119163 98 + 973955 AAAUGGCAUCAGGCACGUGUAAGGGCAACAGCAGGAGCUGUAUAGCAGCAGCAGCAA--GGAUAAUGAUGAAAAUGGUUAAUAUCCGAACGAAAAGAGAG----------- .....((.((..((...(((....)))...))..))(((((......))))).))..--(((((.((((.......)))).)))))..............----------- ( -21.00, z-score = -1.33, R) >droWil1.scaffold_181017 9987 81 + 939937 ------------GUGUGUGCCAGGGCCAGGCACGUAGU-GUA-AGUGGCACUGGCCACUGGAUAAUGAUGAAAACGGUUAAUACUUAUAUGAACG---------------- ------------..((((.((((((((((((((.....-...-.)).)).)))))).)))).((((..(....)..))))))))...........---------------- ( -23.00, z-score = -0.79, R) >consensus ____________AAAUGUCCAACGGUCACGUGUGAAGCAGUA_ACUG___CUGUCCACUGGAUAAUGACGAAAAUGGUUAAUAUCCGUUCGCUUGGAA_AAA_GCGAG_C_ ...........................................................(((((.((((.......)))).)))))......................... ( -6.96 = -6.53 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:23 2011