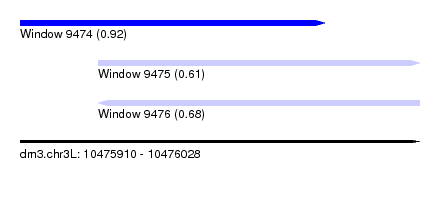
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,475,910 – 10,476,028 |
| Length | 118 |
| Max. P | 0.917819 |

| Location | 10,475,910 – 10,476,000 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 71.09 |
| Shannon entropy | 0.60280 |
| G+C content | 0.44765 |
| Mean single sequence MFE | -10.50 |
| Consensus MFE | -6.75 |
| Energy contribution | -6.84 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10475910 90 + 24543557 AGCAUCCCAAC-UAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACAAACUCACCCAUUUCGCCAUCCCGUUUCCCCGUUAAACAAAG-------- .((........-...(((.....)))(((((((.........)))))))...............))......((((.......))))....-------- ( -7.50, z-score = -1.40, R) >droSim1.chr3L 9872424 89 + 22553184 AGCAUCCCAAC-UAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACAAACCCACCCAUUUCGCCACCCCGUUUACC-GUUAAACAGAG-------- .((........-...(((.....)))(((((((.........)))))))...............))......(((((..-..)))))....-------- ( -9.30, z-score = -1.53, R) >droSec1.super_0 2686591 89 + 21120651 AGCAUCCCAAC-UAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACAAACCCACCCAUUUCGCCACCCCGUUUCCC-GUUAAAUAGAG-------- .((........-...(((.....)))(((((((.........)))))))...............)).............-...........-------- ( -7.40, z-score = -1.08, R) >droEre2.scaffold_4784 10467597 94 + 25762168 AGCAUCCCAAC-UAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACAAACCCACCCACUUCGCCACCCCAUUUCCC-GUUAAACAGGGGGUGG--- .((((......-..........))))(((((((.........)))))))................(((((((.......-.........)))))))--- ( -19.28, z-score = -1.25, R) >droYak2.chr3L 10467495 89 + 24197627 AGCAUCCCAAC-UAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACAAACCCACCCACUUCGCCACCCCAUUUCCC-GUUAAACAGAG-------- .((........-...(((.....)))(((((((.........)))))))...............)).............-...........-------- ( -7.40, z-score = -1.28, R) >droAna3.scaffold_13337 15387698 76 + 23293914 AGCUUCCCGGU-UAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACGAACCCACCUAUUUC-UCUUCCCGUCUGG--------------------- .......(((.-.(((.(((((.((((((((((.........))))))))....))..))))).-.))).))).....--------------------- ( -9.90, z-score = -0.05, R) >dp4.chrXR_group6 8880461 98 + 13314419 AGCAUCCCAACGUAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACGAACACUCCCAUUUCUCAUCAGC-AUCCCCUAAGCUCUCCCUCACCUUUG (((......((....)).....(((((((((((.........)))))))(((.........)))......))-)).......))).............. ( -11.60, z-score = -2.39, R) >droWil1.scaffold_181024 1731893 83 - 1751709 AGCAUCCAAAC-UAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACGAACGAAACGAAAUGAAACCCUCCUCAAAACGUUC--------------- .((((......-..........))))(((((((.........)))))))(((((....((...((.....)).))....)))))--------------- ( -9.39, z-score = -0.38, R) >droVir3.scaffold_13049 16265705 79 + 25233164 AGCAUCCCAAC-UAAGUAAAUAAUGCGUAAAACAACUCGAGCGUUUUACGGUUGCCCACUUCGCACCGCCUCAACCCGCC------------------- .((........-..............(((((((.........)))))))(((.((.......)))))))...........------------------- ( -11.50, z-score = 0.02, R) >droGri2.scaffold_15110 12155338 81 - 24565398 AGCAUCCCAAC-UAAGUAAAUAAUGCGUAAAACAAGUCGAGCGUUUUACGGUUACCCACGCCUCCCACCAACCGUGCCCCGC----------------- .((((......-..............(((((((.........)))))))((((................)))))))).....----------------- ( -10.39, z-score = 0.48, R) >droMoj3.scaffold_6680 4277232 81 - 24764193 AGCAUCCCAAC-UAAGUAAAUAAUGCGUAAAACAAGUCGAGCGUUUUACGGUUGCGUCCUUAGCCCCCUCUCCCUCAGUCAC----------------- .(((.((....-...(((.....)))(((((((.........))))))))).)))...........................----------------- ( -11.80, z-score = -0.71, R) >consensus AGCAUCCCAAC_UAAGUAAAUAAUGCGUAAAACAAGUCAAGCGUUUUACGAACCCACCCAUUUCGCCACCCCGUUUCCC_GUUA_______________ .((((.................))))(((((((.........))))))).................................................. ( -6.75 = -6.84 + 0.09)

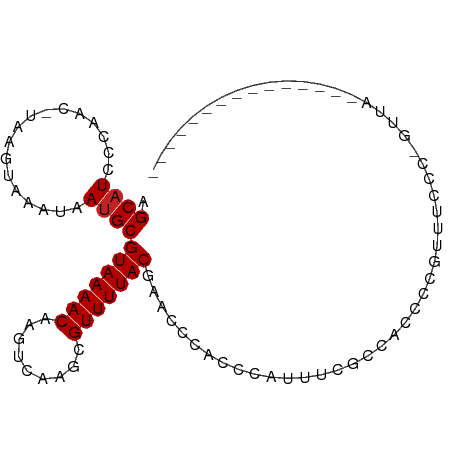
| Location | 10,475,933 – 10,476,028 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 55.86 |
| Shannon entropy | 0.88766 |
| G+C content | 0.51120 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -5.50 |
| Energy contribution | -5.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
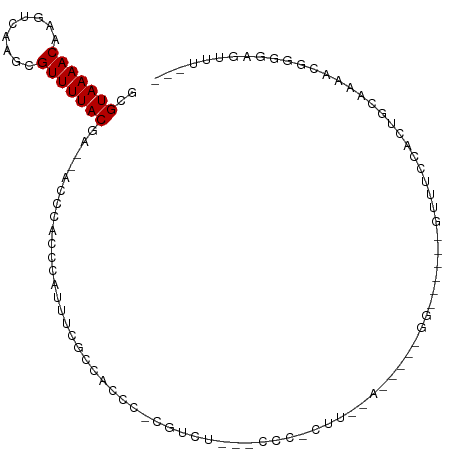
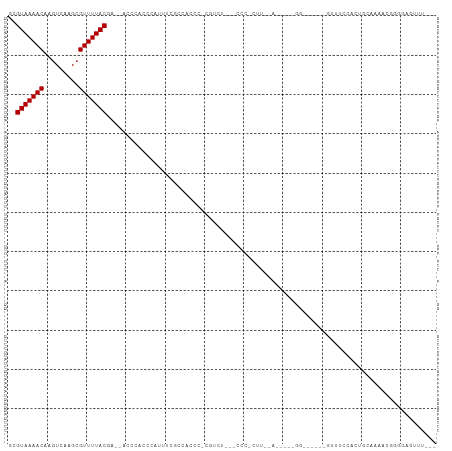
>dm3.chr3L 10475933 95 + 24543557 GCGUAAAACAAGUCAAGCGUUUUACAA--ACUCACCCAUUUCGCCAUCC-CGUUU---CCCCGUUAAACAAAGGGG-----GUUUCCACUGCAAAACGGGGAGUUU--- ..(((((((.........)))))))((--((((.(((.(((.((.....-....(---((((..........))))-----)........)).))).)))))))))--- ( -26.43, z-score = -2.31, R) >droSim1.chr3L 9872447 94 + 22553184 GCGUAAAACAAGUCAAGCGUUUUACAA--ACCCACCCAUUUCGCCACCC-CGUUU---ACC-GUUAAACAGAGGGG-----GUUUCCACUGCAAAACGGGGAGUUU--- ..(((((((.........)))))))((--((.(.(((.........(((-(((((---(..-..)))))...))))-----((((........)))))))).))))--- ( -21.20, z-score = 0.03, R) >droSec1.super_0 2686614 94 + 21120651 GCGUAAAACAAGUCAAGCGUUUUACAA--ACCCACCCAUUUCGCCACCC-CGUUU---CCC-GUUAAAUAGAGGGG-----GUUUCCACUGCAAAACGGGGAGUUU--- ..(((((((.........)))))))..--.................(((-(((((---(((-.((.....)).)))-----((.......)).)))))))).....--- ( -23.00, z-score = -0.60, R) >droEre2.scaffold_4784 10467620 99 + 25762168 GCGUAAAACAAGUCAAGCGUUUUACAA--ACCCACCCACUUCGCCACCC-CAUUU---CCC-GUUAAACAGGGGGUGGGGUGUUUUCACUGCAAAAUGGGGAGUUU--- ..(((((((.........)))))))((--((.(.((((.((.(((((((-(((..---((.-(.....).))..))))))))........)).)).))))).))))--- ( -34.40, z-score = -2.25, R) >droYak2.chr3L 10467518 94 + 24197627 GCGUAAAACAAGUCAAGCGUUUUACAA--ACCCACCCACUUCGCCACCC-CAUUU---CCC-GUUAAACAGAGGGG-----GUUUUCACUGCAAAAUGGGGAGUUU--- ..(((((((.........)))))))..--.................(((-(((((---(((-.((.....)).)))-----((.......)).)))))))).....--- ( -21.60, z-score = -0.32, R) >droAna3.scaffold_13337 15387721 81 + 23293914 GCGUAAAACAAGUCAAGCGUUUUACGA--ACCCACCUAUUUCUCU-UCC-CGUCU---GGUGAU------------------UCUCCACUUCCUAGCUGGGAAUCC--- .((((((((.........)))))))).--...............(-(((-((.((---((.((.------------------........)))))).))))))...--- ( -19.40, z-score = -2.66, R) >dp4.chrXR_group6 8880485 109 + 13314419 GCGUAAAACAAGUCAAGCGUUUUACGAACACUCCCAUUUCUCAUCAGCAUCCCCUAAGCUCUCCCUCACCUUUGGUGGGUCGGCUCUCUUCCCCCGCCCCACCCCCGGC .((((((((.........))))))))...................(((.........)))........((...((((((.(((..........))).))))))...)). ( -23.60, z-score = -2.08, R) >droWil1.scaffold_181024 1731916 83 - 1751709 GCGUAAAACAAGUCAAGCGUUUUACGAACGAAACGAAAUGAAACCCUCCUCAAAAC-GUUCACCG-----------------UCUCCAUCUCCAUUUCCAU-------- .((((((((.........)))))))).(((.((((...(((........)))...)-)))...))-----------------)..................-------- ( -10.50, z-score = -1.22, R) >droVir3.scaffold_13049 16265728 84 + 25233164 GCGUAAAACAACUCGAGCGUUUUACGGUUG---CCCACUUCGCACCGCCUCAACCC-GCCCUCCU-----------------UCUCU-CACCAUCCUAUAGAGCUC--- (((((((((.........)))))))(((.(---(.......)))))))........-........-----------------.((((-...........))))...--- ( -11.80, z-score = -0.63, R) >droGri2.scaffold_15110 12155361 85 - 24565398 GCGUAAAACAAGUCGAGCGUUUUACGGUUACCCACGCCUCCCACCAACCGUGCCC---CGCCCCU-----------------UUAGUCCAGCAACGCUAAACACC---- ...........((..((((((.(((((((................)))))))...---.((....-----------------........))))))))..))...---- ( -14.29, z-score = -0.77, R) >droMoj3.scaffold_6680 4277255 81 - 24764193 GCGUAAAACAAGUCGAGCGUUUUACGGUUG---CGUCCUUAGCCCCCUCUCC-CUC-AGUCACCU-----------------CCUCC-UCUCAGC--ACACAGCUC--- (((((((((.........)))))))(((((---......)))))........-...-........-----------------.....-.....))--.........--- ( -11.20, z-score = -0.19, R) >consensus GCGUAAAACAAGUCAAGCGUUUUACGA__ACCCACCCAUUUCGCCACCC_CGUCU___CCC_CUU__A_____GG______GUUUCCACUGCAAAACGGGGAGUUU___ ..(((((((.........))))))).................................................................................... ( -5.50 = -5.50 + 0.00)
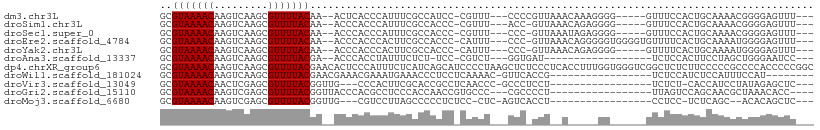
| Location | 10,475,933 – 10,476,028 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 55.86 |
| Shannon entropy | 0.88766 |
| G+C content | 0.51120 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -6.71 |
| Energy contribution | -6.46 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
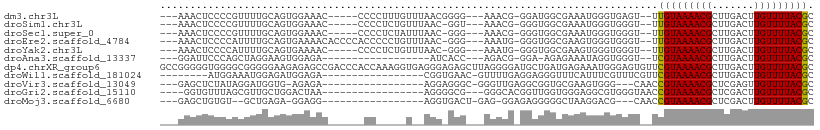
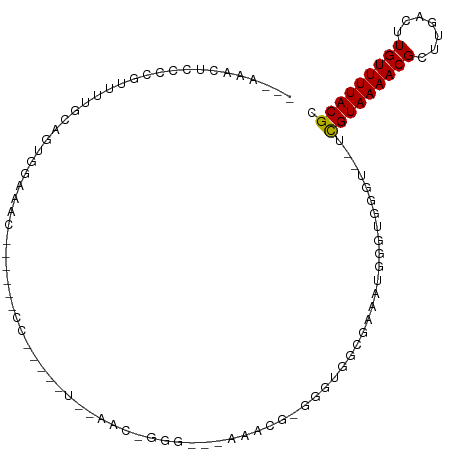
>dm3.chr3L 10475933 95 - 24543557 ---AAACUCCCCGUUUUGCAGUGGAAAC-----CCCCUUUGUUUAACGGGG---AAACG-GGAUGGCGAAAUGGGUGAGU--UUGUAAAACGCUUGACUUGUUUUACGC ---((((((((((((((((...(....)-----((((..........))))---.....-.....)))))))))).))))--))((((((((.......)))))))).. ( -35.20, z-score = -3.17, R) >droSim1.chr3L 9872447 94 - 22553184 ---AAACUCCCCGUUUUGCAGUGGAAAC-----CCCCUCUGUUUAAC-GGU---AAACG-GGGUGGCGAAAUGGGUGGGU--UUGUAAAACGCUUGACUUGUUUUACGC ---((((((((((((((((...(....)-----..((((((((((..-..)---)))))-))).))))))))))).))))--))((((((((.......)))))))).. ( -36.30, z-score = -3.04, R) >droSec1.super_0 2686614 94 - 21120651 ---AAACUCCCCGUUUUGCAGUGGAAAC-----CCCCUCUAUUUAAC-GGG---AAACG-GGGUGGCGAAAUGGGUGGGU--UUGUAAAACGCUUGACUUGUUUUACGC ---((((((((((((((((.......((-----(((.(((.......-.))---)...)-)))).)))))))))).))))--))((((((((.......)))))))).. ( -34.80, z-score = -2.59, R) >droEre2.scaffold_4784 10467620 99 - 25762168 ---AAACUCCCCAUUUUGCAGUGAAAACACCCCACCCCCUGUUUAAC-GGG---AAAUG-GGGUGGCGAAGUGGGUGGGU--UUGUAAAACGCUUGACUUGUUUUACGC ---((((((((((((((((.((....))((((((...((((.....)-)))---...))-)))).)))))))))).))))--))((((((((.......)))))))).. ( -44.40, z-score = -4.49, R) >droYak2.chr3L 10467518 94 - 24197627 ---AAACUCCCCAUUUUGCAGUGAAAAC-----CCCCUCUGUUUAAC-GGG---AAAUG-GGGUGGCGAAGUGGGUGGGU--UUGUAAAACGCUUGACUUGUUUUACGC ---((((((((((((((((.......((-----(((.((((.....)-)))---....)-)))).)))))))))).))))--))((((((((.......)))))))).. ( -36.40, z-score = -3.33, R) >droAna3.scaffold_13337 15387721 81 - 23293914 ---GGAUUCCCAGCUAGGAAGUGGAGA------------------AUCACC---AGACG-GGA-AGAGAAAUAGGUGGGU--UCGUAAAACGCUUGACUUGUUUUACGC ---.((((((((.((....))))).))------------------))).((---....)-)..-.(.(((((((((.((.--.((.....)).)).))))))))).).. ( -17.60, z-score = 0.21, R) >dp4.chrXR_group6 8880485 109 - 13314419 GCCGGGGGUGGGGCGGGGGAAGAGAGCCGACCCACCAAAGGUGAGGGAGAGCUUAGGGGAUGCUGAUGAGAAAUGGGAGUGUUCGUAAAACGCUUGACUUGUUUUACGC (((...((((((.(((..........))).))))))...)))........((.....(((((((..(.(....).).)))))))((((((((.......)))))))))) ( -33.00, z-score = -0.77, R) >droWil1.scaffold_181024 1731916 83 + 1751709 --------AUGGAAAUGGAGAUGGAGA-----------------CGGUGAAC-GUUUUGAGGAGGGUUUCAUUUCGUUUCGUUCGUAAAACGCUUGACUUGUUUUACGC --------...(((((((((.(.((((-----------------((.....)-))))).)......)))))))))........(((((((((.......))))))))). ( -20.60, z-score = -0.78, R) >droVir3.scaffold_13049 16265728 84 - 25233164 ---GAGCUCUAUAGGAUGGUG-AGAGA-----------------AGGAGGGC-GGGUUGAGGCGGUGCGAAGUGGG---CAACCGUAAAACGCUCGAGUUGUUUUACGC ---...............(((-(((.(-----------------(...((((-(..((...(((((((.......)---).))))).)).)))))...)).)))))).. ( -22.80, z-score = -0.65, R) >droGri2.scaffold_15110 12155361 85 + 24565398 ----GGUGUUUAGCGUUGCUGGACUAA-----------------AGGGGCG---GGGCACGGUUGGUGGGAGGCGUGGGUAACCGUAAAACGCUCGACUUGUUUUACGC ----...(((((((...)))))))...-----------------..(((((---....(((((((.(.(......).).)))))))....))))).............. ( -22.90, z-score = 0.65, R) >droMoj3.scaffold_6680 4277255 81 + 24764193 ---GAGCUGUGU--GCUGAGA-GGAGG-----------------AGGUGACU-GAG-GGAGAGGGGGCUAAGGACG---CAACCGUAAAACGCUCGACUUGUUUUACGC ---.......((--(.(((((-.(((.-----------------((....))-(((-.(...((..((.......)---)..))......).)))..))).)))))))) ( -18.90, z-score = 0.80, R) >consensus ___AAACUCCCCGUUUUGCAGUGGAAAC______CC_____U__AAC_GGG___AAACG_GGGUGGCGAAAUGGGUGGGU__UCGUAAAACGCUUGACUUGUUUUACGC ...................................................................................(((((((((.......))))))))). ( -6.71 = -6.46 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:20 2011