| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,446,017 – 10,446,107 |
| Length | 90 |
| Max. P | 0.506507 |

| Location | 10,446,017 – 10,446,107 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.75 |
| Shannon entropy | 0.45845 |
| G+C content | 0.49934 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -9.47 |
| Energy contribution | -9.89 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
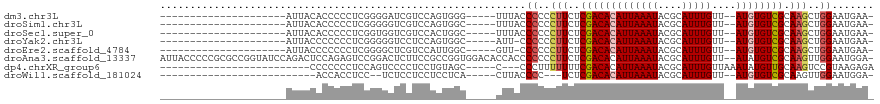
>dm3.chr3L 10446017 90 - 24543557 ---------------------AUUACACCCCCUCGGGGAUCGUCCAGUGGG-----UUUACCCCCCUUCUCGACACAUUAAAUACGCAUUUGUU--AUGUGUCGCAAGCUGGAAUGAA- ---------------------......((((...)))).((((((((((((-----....))).......(((((((((((((....)))))..--))))))))...))))).)))).- ( -29.60, z-score = -2.79, R) >droSim1.chr3L 9841254 90 - 22553184 ---------------------AUUACACCCCCUCGGGGGUCGUCCAGUGGC-----UUUACCCCCCUUCUCGACACAUUAAAUACGCAUUUGUU--AUGUGUCGCAAGCUGGAAUGAA- ---------------------......((((....))))(((((((((((.-----....))........(((((((((((((....)))))..--))))))))...))))).)))).- ( -27.90, z-score = -2.23, R) >droSec1.super_0 2655934 90 - 21120651 ---------------------AUUACACCCCCUCGGUGGUCGUCCACUGGC-----UUUACCCCCCUUCUCGACACAUUAAAUACGCAUUUGUU--AUGUGUCGCAAGCUGGAAUGAA- ---------------------...........(((((((....))))))).-----.....((..(((..(((((((((((((....)))))..--)))))))).)))..))......- ( -21.40, z-score = -1.28, R) >droYak2.chr3L 10436039 89 - 24197627 ---------------------AUUACCCCCCCUCGGGGGUCCUCCAGUGGC-----AUU-CCCCCCUUCUCGACACAUUAAAUACGCAUUUGUU--AUGUGUCGCAAGCUGGAAUGAA- ---------------------...((((((....))))))((......))(-----(((-((...(((..(((((((((((((....)))))..--)))))))).)))..))))))..- ( -29.00, z-score = -2.90, R) >droEre2.scaffold_4784 10436171 89 - 25762168 ---------------------AUUACCCCCCCUCGGGGCUCGUCCAUUGGC-----GUU-CCCCCCUUCUCGACACAUUAAAUACGCAUUUGUU--AUGUGUCGCAAGCUGGAAUGAA- ---------------------.............((((..((((....)))-----)..-))))((..(((((((((((((((....)))))..--))))))))..))..))......- ( -24.80, z-score = -1.85, R) >droAna3.scaffold_13337 15356983 116 - 23293914 AUUACCCCCGCGCCGGUAUCCAGACUCCAGAGUCCGGACUCUUCCGCCGGUGGACACCACCCCCCCUUCUCGACACAUUAAAUACGCAUUUGUU--AUAUGUCGCAAGUUGGAAUGGA- ....((((.(((..((..(((.((((....)))).)))..))..))).)).))...(((..((..(((..(((((...(((((....)))))..--...))))).)))..))..))).- ( -30.70, z-score = -0.97, R) >dp4.chrXR_group6 4467841 86 + 13314419 -------------------------CCCCCCCUCCAGUCCCCUCCUGUAGC-----C---CCCUUUUUUUCGACACAUUAAAUACGCAUUUGUUAAAUAUGUUGCAAGUCCGUAAGAGA -------------------------......((((((.......)))....-----.---......................((((.((((((..........)))))).)))).))). ( -9.70, z-score = -1.40, R) >droWil1.scaffold_181024 1674022 80 + 1751709 --------------------------ACCACCUCC--UCUCCUCCUCCUCA-----CUUACCCC---UCUCGACACAUUAAAUACGCAUUUGUU--AUGUGUCGCAAGUUGGAAUGGA- --------------------------.........--..(((...(((..(-----(((.....---...(((((((((((((....)))))..--)))))))).)))).)))..)))- ( -14.60, z-score = -2.16, R) >consensus _____________________AUUACACCCCCUCGGGGCUCGUCCAGUGGC_____CUUACCCCCCUUCUCGACACAUUAAAUACGCAUUUGUU__AUGUGUCGCAAGCUGGAAUGAA_ .............................................................((..(((..((((((((..((((......))))..)))))))).)))..))....... ( -9.47 = -9.89 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:18 2011