| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,761,203 – 4,761,306 |
| Length | 103 |
| Max. P | 0.847650 |

| Location | 4,761,203 – 4,761,306 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.58 |
| Shannon entropy | 0.63394 |
| G+C content | 0.43597 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -8.32 |
| Energy contribution | -7.87 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4761203 103 + 23011544 ---CUGAUUCCCAACCCGAUUCGCAUUGCAAUUGCAAAGUAAAUGCUUUAAUGCGCUUAUCCUUUUAGCG--UCGGUUGGGCAUUCGUUCAGACUACC--UAACCGAAUG------- ---......(((((((.(((..(((((((....))(((((....))))))))))(((.........))))--)))))))))((((((...((.....)--)...))))))------- ( -28.00, z-score = -2.73, R) >droSim1.chr2L 4630530 103 + 22036055 ---CUGAUUCCCAACCCGAUUCGCAUUGCAAUUGCAAAGUAAAUGCUUUAAUGCGCUUAUCCUUUUAGCG--UCGGUUGGGCAUUCGUUCAGACUACC--UAACCGAAUG------- ---......(((((((.(((..(((((((....))(((((....))))))))))(((.........))))--)))))))))((((((...((.....)--)...))))))------- ( -28.00, z-score = -2.73, R) >droSec1.super_5 2857595 103 + 5866729 ---CUGAUUCCCAACCCGAUUCGCAUUGCAAUUGCAAAGUAAAUGCUUUAAUGCGCUUAUCCUUUUAGCG--UCGGUUGGGCAUUCGUUCAGACUACC--UAACCGAAUG------- ---......(((((((.(((..(((((((....))(((((....))))))))))(((.........))))--)))))))))((((((...((.....)--)...))))))------- ( -28.00, z-score = -2.73, R) >droYak2.chr2L 4769090 89 + 22324452 -----------------GAUUCGCAUUGCAAUUGCAAAGUAAAUGCUUUAAUGCGCUUAUCCUUUUAGCG--UCGGUUGGGCAUUCGCUCAGACUACC--UAACCGAAUG------- -----------------(((.((((((((....))(((((....)))))))))))...))).........--(((((((((...((.....))...))--)))))))...------- ( -24.80, z-score = -2.11, R) >droEre2.scaffold_4929 4836660 91 + 26641161 ---------------CUGAUUCGCAUUGCAAUCGCAAAGUAAAUGCUUUAAUGCGCUUAUCCUUUUAGCG--UCGGUUGGGCAUUCGUUCAGACUACC--UAACCGAAUG------- ---------------..(((.((((((((....))(((((....)))))))))))...))).........--(((((((((...((.....))...))--)))))))...------- ( -24.50, z-score = -2.03, R) >droAna3.scaffold_12943 1962277 112 - 5039921 ---UUGAUUCCCGCCCCGAUUCCCAUUGCAAUUGCAAAGUAAAUGCCCUAAUGCGCUUAUCCUUUUAGCG--CCAGUUGGGCAUUUGUUCAGCCACCCAUUUCCCGAGCCCAUUUCA ---.(((.....((..((.......((((....))))..(((((((((....(((((.........))))--).....))))))))).................)).)).....))) ( -25.60, z-score = -2.29, R) >dp4.chr4_group3 620926 91 - 11692001 ---GUGAUUCUCUGCCUGAUUCGCAUUCCAAUUGCAAAGUAAAUGCCUUAAUGUGCUUAUCCUUUUAGCA--ACGGUUGGGCAUUCGCUUAGAGUU--------------------- ---......((((.........(((.......))).(((..(((((((...((((((.........))).--)))...)))))))..)))))))..--------------------- ( -23.90, z-score = -1.71, R) >droPer1.super_8 1675321 91 - 3966273 ---GUGAUUCUCUGCCUGAUUCGCAUUCCAAUUGCAAAGUAAAUGCCUUAAUGUGCUUAUCCUUUUAGCA--ACGGUUGGGCAUUCGCUUAGAGUU--------------------- ---......((((.........(((.......))).(((..(((((((...((((((.........))).--)))...)))))))..)))))))..--------------------- ( -23.90, z-score = -1.71, R) >droWil1.scaffold_180703 1503028 110 - 3946847 UUGUGGCUGGCACUCGGCAUUCCAAUUGCAAUUGCAAAGUAAAUGCCUUAAUGCGCUUAUCCUUUUAGCGAGUCAGUUGAGCAUUCGUUUAUACUUCACCCCAGGUUUUU------- ..(((((((((....((((((....((((....))))....))))))......((((.........)))).))))))(((((....))))).....)))...........------- ( -26.30, z-score = -0.56, R) >droVir3.scaffold_12963 8765794 89 - 20206255 -----------------UCCUCUGAUUCGCAUUGCAAAGUAAAUGCCUUAAUGCGCUUAUCCUUUUAGCG--CCAGUUAAGUAUUCGGUU-UGCUUAUAUACACGCCCU-------- -----------------................(((((.(.(((((.((((((((((.........))))--)..)))))))))).).))-)))...............-------- ( -17.90, z-score = -1.10, R) >droMoj3.scaffold_6500 12157834 83 - 32352404 ----------------UUGCCCAGAUUCGCAUCGCAAAGUAAAUGCCUUAAUGCGCUUAUCCUUUUAGCG--UCAGUUGAGCAUUCGAUU-UACUUAGACCC--------------- ----------------.(((........))).....((((((((((.((((((((((.........))))--)..))))))))).....)-)))))......--------------- ( -14.50, z-score = -0.38, R) >droGri2.scaffold_15126 3307227 90 - 8399593 ----------------UCCGUUUGAAUCGCAAUCCAAAGUAAAUGCCCUAAUGCGCUUAUCCUUUUAGCG--UCAGUUGAGCAUUCGAUU-UACUUAUACCACCAACCA-------- ----------------......(((((((............(((((.(...((((((.........))))--.))...).))))))))))-))................-------- ( -14.00, z-score = -1.45, R) >consensus ____UGAUUC_C__CCUGAUUCGCAUUGCAAUUGCAAAGUAAAUGCCUUAAUGCGCUUAUCCUUUUAGCG__UCGGUUGGGCAUUCGUUCAGACUUCC__UAACCGAAU________ .....................................((..(((((.(((((.((((.........)))).....))))))))))..))............................ ( -8.32 = -7.87 + -0.46)


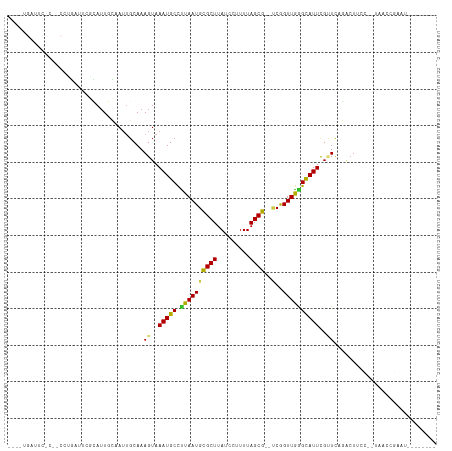
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:22 2011