| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,427,384 – 10,427,562 |
| Length | 178 |
| Max. P | 0.765676 |

| Location | 10,427,384 – 10,427,562 |
|---|---|
| Length | 178 |
| Sequences | 5 |
| Columns | 179 |
| Reading direction | forward |
| Mean pairwise identity | 71.88 |
| Shannon entropy | 0.49175 |
| G+C content | 0.32478 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -16.81 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
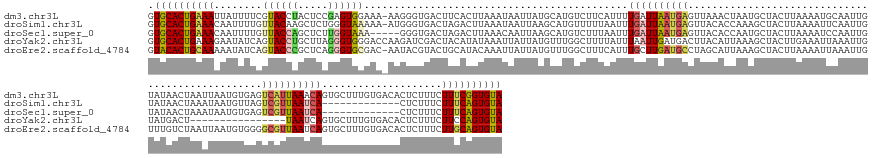
>dm3.chr3L 10427384 178 + 24543557 GUGCACUGAAAUUAUUUUCGUACCUACUCCGAGUGGAAA-AAGGGUGACUUCACUUAAAUAAUUAUGCAUGUCUUCAUUUUGAUUAAUGAGUUAAACUAAUGCUACUUAAAAUGCAAUUGUAUAACUAAUUAAUGUGAGUCAUUAAACAGUGCUUUGUGACACUCUUUCUUUCGGUGUA .((((((((((........((....))...(((((....-...(((((((.(((((((.((.(((((((....((((((......)))))).........(((..........)))..))))))).)).)))).))))))))))..((((....))))..)))))....)))))))))) ( -40.20, z-score = -2.14, R) >droSim1.chr3L 9822796 165 + 22553184 GUGCACUGAAACAAUUUUGUUACAAGCUCUGGGUAAAAA-AUGGGUGACUAGACUUAAAUAAUUAAGCAUGUUUUUAAUUUGAUUAAUGAGUUACACCAAAGCUACUUAAAAUUCAAUUGUAUAACUAAAUAAUGUUAGUCGUUAAUCA-------------CUCUUUCUUUCAGUGUA .(((((((((((((((..(((...((((.(((((.....-.(((....)))((((((..((((((((...........)))))))).)))))))).))).))))......)))..)))))).........(((((.....)))))....-------------........))))))))) ( -32.60, z-score = -1.24, R) >droSec1.super_0 2637464 161 + 21120651 GUGCACUGAAACAAUUUUGUUACCAGCUCUUGGUAAA-----GGGUGACUAGACUUAAACAAUUAAGCAUGUCUUUAAUUUGAUUAAUGAGUUACACCAAUGCUACUUAAAAUCCAAUUGUAUAACUAAAUAAUGUGAGUCGUUAAUCA-------------CUCUUUCUUUCAGUGUA .((((((((((.........((((((...))))))((-----(((((((..(.(((((....))))))..)))........(((((((((.(((((.((((...............))))(((......))).))))).))))))))))-------------)))))..)))))))))) ( -37.36, z-score = -2.38, R) >droYak2.chr3L 10417086 163 + 24197627 GUGCACUGAAAGAAUAUCAGUACCUGCUUAGGGUGGGACCAAGAUCGACUACAUAUAAAUUAUUAUGUUUGGCUUUUAUUUAAUUGAUGACUUACAUUAAAGCUACUUGAAAUUAAAUUGUAUGACU----------------UAAUCAGUGCUUUGUGACACUCUUUCUUCCAGUGUA .(((((((.((((((((((((.((..(.....)..))...(((.((((..(((((........)))))))))))).......))))))).....(((.(((((.(((.((..((((.((....)).)----------------)))))))))))))))).......))))).))))))) ( -34.30, z-score = -0.71, R) >droEre2.scaffold_4784 10417119 178 + 25762168 GUACACUGCAAAAAUAUCAGUACCCGCUCAGGGUGCGAC-AAUACGUACUGCAUACAAAUUAUUAUGUUUGGCUUUCAUUUGCUUGAUGCCUAGCAUUAAAGCUACUUAAAAUUAAAUUGUUUGUCUAAUUAAUGUGGGGCGUUAAUCAGUGCUUUGUGACACUCUUUCUUGCAGUGUA .((((((((((........((((((.....)))))).((-((...((((((....(((((......)))))((........))(((((((((.(((((((.........((((......))))......))))))).))))))))).)))))).))))...........)))))))))) ( -52.66, z-score = -3.20, R) >consensus GUGCACUGAAACAAUUUUAGUACCAGCUCAGGGUGAAAA_AAGGGUGACUACACUUAAAUAAUUAUGCAUGUCUUUAAUUUGAUUAAUGAGUUACACUAAAGCUACUUAAAAUUCAAUUGUAUAACUAAAUAAUGUGAGUCGUUAAUCAGUGCUUUGUGACACUCUUUCUUUCAGUGUA .((((((((((........((((((.....))))))............................................((((((((((.................................................))))))))))....................)))))))))) (-16.81 = -17.53 + 0.72)

| Location | 10,427,384 – 10,427,562 |
|---|---|
| Length | 178 |
| Sequences | 5 |
| Columns | 179 |
| Reading direction | reverse |
| Mean pairwise identity | 71.88 |
| Shannon entropy | 0.49175 |
| G+C content | 0.32478 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -12.92 |
| Energy contribution | -13.38 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
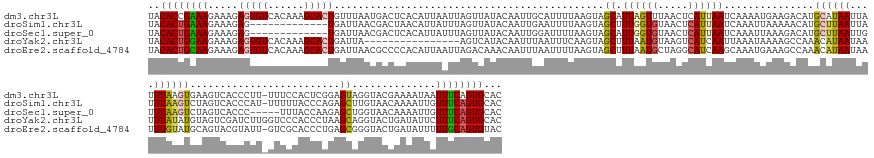
>dm3.chr3L 10427384 178 - 24543557 UACACCGAAAGAAAGAGUGUCACAAAGCACUGUUUAAUGACUCACAUUAAUUAGUUAUACAAUUGCAUUUUAAGUAGCAUUAGUUUAACUCAUUAAUCAAAAUGAAGACAUGCAUAAUUAUUUAAGUGAAGUCACCCUU-UUUCCACUCGGAGUAGGUACGAAAAUAAUUUCAGUGCAC (((.((((...(((.(((((......))))).)))..((((((((.((((.(((((((.....(((.......)))((((..((((...(((((......)))))))))))))))))))).))))))).))))).....-.......)))).))).((((((((....)))).)))).. ( -39.70, z-score = -3.03, R) >droSim1.chr3L 9822796 165 - 22553184 UACACUGAAAGAAAGAG-------------UGAUUAACGACUAACAUUAUUUAGUUAUACAAUUGAAUUUUAAGUAGCUUUGGUGUAACUCAUUAAUCAAAUUAAAAACAUGCUUAAUUAUUUAAGUCUAGUCACCCAU-UUUUUACCCAGAGCUUGUAACAAAAUUGUUUCAGUGCAC ..((((((((.......-------------........((((((......))))))...(((((.....(((...((((((((.((((....(((((...)))))......((((((....))))))............-...))))))))))))..)))...)))))))))))))... ( -31.40, z-score = -1.53, R) >droSec1.super_0 2637464 161 - 21120651 UACACUGAAAGAAAGAG-------------UGAUUAACGACUCACAUUAUUUAGUUAUACAAUUGGAUUUUAAGUAGCAUUGGUGUAACUCAUUAAUCAAAUUAAAGACAUGCUUAAUUGUUUAAGUCUAGUCACCC-----UUUACCAAGAGCUGGUAACAAAAUUGUUUCAGUGCAC ..((((((((.((((.(-------------((((((..((((...((((...((((((((...((.((.....))..))...))))))))...))))........(((((........))))).))))))))))).)-----)((((((.....)))))).....)).))))))))... ( -36.00, z-score = -1.82, R) >droYak2.chr3L 10417086 163 - 24197627 UACACUGGAAGAAAGAGUGUCACAAAGCACUGAUUA----------------AGUCAUACAAUUUAAUUUCAAGUAGCUUUAAUGUAAGUCAUCAAUUAAAUAAAAGCCAAACAUAAUAAUUUAUAUGUAGUCGAUCUUGGUCCCACCCUAAGCAGGUACUGAUAUUCUUUCAGUGCAC ..((((((((((....((((((.(((((((((((((----------------(((......))))))))...))).))))).........................(((..(((((........))))).......(((((.......)))))..)))..))))))))))))))))... ( -32.50, z-score = -1.53, R) >droEre2.scaffold_4784 10417119 178 - 25762168 UACACUGCAAGAAAGAGUGUCACAAAGCACUGAUUAACGCCCCACAUUAAUUAGACAAACAAUUUAAUUUUAAGUAGCUUUAAUGCUAGGCAUCAAGCAAAUGAAAGCCAAACAUAAUAAUUUGUAUGCAGUACGUAUU-GUCGCACCCUGAGCGGGUACUGAUAUUUUUGCAGUGUAC (((((((((((((..(((((......))))).((((..((((...........(((((((((.(((........((((......))))(((.(((......)))..)))........))).)))).(((.....)))))-)))((.......))))))..)))).))))))))))))). ( -46.20, z-score = -2.51, R) >consensus UACACUGAAAGAAAGAGUGUCACAAAGCACUGAUUAACGACUCACAUUAAUUAGUUAUACAAUUGAAUUUUAAGUAGCUUUAGUGUAACUCAUUAAUCAAAUUAAAGACAUGCAUAAUUAUUUAAGUGUAGUCACCCUU_GUUCCACCCAGAGCAGGUACCAAAAUUGUUUCAGUGCAC ..((((((((.....(((((......))))).............................................((.((((((.....))))))...............((((((....)))))).........................))..............))))))))... (-12.92 = -13.38 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:13 2011